Abstract
Saliva is a body fluid of a unique composition devoted to protect the mouth cavity and the digestive tract. Our high performance liquid chromatography (HPLC)-electrospray ionization-MS analysis of the acidic soluble fraction of saliva from preterm human newborn surprisingly revealed more than 40 protein masses often undetected in adult saliva. We were able to identify the following proteins: stefin A and stefin B, S100A7 (two isoforms), S100A8, S100A9 (four isoforms), S100A11, S100A12, small proline-rich protein 3 (two isoforms), lysozyme C, thymosins β4 and β10, antileukoproteinase, histone H1c, and α and γ globins. The average mass value reported in international data banks was often incongruent with our experimental results mostly because of post-translational modifications of the proteins, e.g. acetylation of the N-terminal residue. A quantitative label-free MS analysis showed protein levels altered in relation to the postconceptional age and suggested coordinate and hierarchical functions for these proteins during development. In summary, this study shows for the first time that analysis of these proteins in saliva of preterm newborns might represent a noninvasive way to obtain precious information of the molecular mechanisms of development of human fetal oral structures.
Saliva is a body fluid of a very complex and specific composition devoted to the protection and well-being of the oral cavity and, because it is swallowed, of the digestive tract (1). Protection is ensured by organic and inorganic solutes and specific peptides and proteins, such as acidic and basic proline-rich proteins, α-amylases, salivary cystatins, histatins, and statherin (2–5). In a previous study (6), we have established that some salivary proteins and peptides reach the levels typically observed in the adult around 18 years of age. Encouraged by the noninvasive specimen collection, we explored the salivary protein composition of at-term and preterm newborns, in order to establish the starting point of the secretion of the proteins and peptides specific of saliva. Our first study (7) showed that acidic proline-rich proteins secretion started, although at very low levels, at 7 months of postconceptional age. At this age the level of phosphorylation of these proteins was low and it increased reaching a value comparable with that of adults at about one year of age, in concomitance with the beginning of deciduous dentition. Other deep differences between human and preterm saliva were however evident. Highly abundant protein masses detected in preterm saliva were undetectable (at the sensitivity level of our MS apparatus) or at very low level in adult saliva. In a previous study (8) we identified, by different MS approaches, thymosin β4 (Tβ4) and thymosin β10 (Tβ10) in preterm newborn saliva and established by immunohistochemistry their presence in fetal salivary glands. This finding let us to suppose that in preterm newborns these peptides derived from gland secretion (8) whereas the low levels in adult saliva were mainly of crevicular fluid origin (9). In another study (10) we were able to elucidate the structure of two isoforms of the small proline-rich protein 3 (SPRR3, or cornifin β)1 detectable only in preterm saliva.
The present study extends our previous findings and reports for the first time the identification of other proteins in the acidic soluble fraction of whole saliva of human preterm newborns. This goal could only be reached by using a top-down proteomic platform based on high resolution ESI-MS measurements following chromatographic separation of intact proteins.
The identification of 20 components was achieved by different chemical and enzymatic treatments coupled to low- and high-resolution MS and mainly confirmed by high-resolution tandem MS (MS/MS) experiments performed on intact proteins. Moreover, the relative amounts of these proteins were determined in 61 different samples of eight preterm subjects in order to evaluate concentration trends as a function of postconceptional age. The relative amount of proteins in whole saliva of preterm newborn was also compared with whole saliva of at-term newborns and adults.
EXPERIMENTAL PROCEDURES
Ethics Statements
The study protocol and written consent forms were approved by both the Pediatric Department Ethics Committee and by the Medical Ethics Committee of the Faculty of Medicine of the Catholic University of Rome (according to the instructions of the Declaration of Helsinki). Full written consent forms were obtained from adult donors and from the parents of the newborns and all rules were respected.
Reagents and Apparatus
All common chemicals and reagents were of analytical grade and were purchased from Carlo Erba, (Milan, Italy), Merck (Darmstadt, Germany), Sigma Aldrich (St. Louis, MO), and Pierce Biotechnology (Rockford, IL). Low-resolution HPLC-ESI-MS measurements were carried out by a Surveyor HPLC system (ThermoFisher, San Jose, CA) connected by a T splitter to a photodiode-array detector and an LCQ Deca XP Plus mass spectrometer (ThermoFisher). The chromatographic column was a Vydac (Hesperia, CA) C8 with 5-μm particle diameter (column dimensions 150 × 2.1 mm). High-resolution HPLC-ESI-MS/MS experiments were carried out by an Ultimate 3000 Nano/Micro HPLC apparatus (Dionex, Sunnyvale, CA) equipped with a FLM-3000-Flow manager module and coupled to an LTQ Orbitrap XL apparatus (ThermoFisher). A Dionex C18 column (3-μm particle diameter; column dimension 300 μm i.d. ×15 cm) or a Zorbax 300 SB-C8 (3.5-μm particle diameter; column dimension 1 mm i.d. ×15 cm) were used as chromatographic columns. Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF)-MS was an Autoflex Brucker Daltonics (Billerica, MA) apparatus.
Subjects Enrolled, Sample Collection and Treatment
Eight newborns (five females, three males) with birth weight between 500 g and 1250 g and 193–217 days of postconceptional age (27–31 weeks), admitted to the Neonatal Intensive Care Unit of the Faculty of Medicine of the Catholic University were enrolled for this study. Infants with major congenital malformations or prenatal infections were excluded from the study. Sample collection was performed on the same preterm newborn during several weeks following birth at established time intervals (1 or 2 weeks). If possible, analysis was also performed following discharge from the neonatal unit during the periodical check visits up to 1 year follow-up. For ethical reasons, saliva was only collected when sample collection caused no stress to the newborn. In this way we were able to collect and analyze 61 saliva specimens from newborns with postconceptional age ranging between 194 and 545 days.
Moreover, saliva from four at term infants (two females, two males) born after uncomplicated pregnancies and vaginal delivery and admitted to Policlinico “A. Gemelli” nursery, was studied. Gestational age and birth weight (Mean ± S.D.) were 272 ± 7 days (38 ± 1 week) and 3280 ± 150 g, respectively. These four newborns had no clinical problems, therefore they were discharged after 3 days of breast-feeding.
Resting whole saliva was also collected with a soft plastic aspirator at the base of the tongue from 20 informed adult volunteers (42 ± 18 years old, 10 males, 10 females).
After collection, salivary samples were immediately mixed with an equal volume of 0.2% aqueous trifluoroacetic acid (TFA) (v/v) in ice bath. After stirring, the acidic supernatant was immediately centrifuged at 9000 × g for 3 min to remove precipitate and the acidic clear solution was either immediately analyzed by HPLC-ESI-MS (100 μl, corresponding to 50 μl of whole saliva) or stored at −80 °C.
Reversed Phase (RP)-HPLC-ESI-MS Analysis
The following solutions were utilized in low-resolution ESI-MS chromatographic separations: (eluent A) 0.056% aqueous TFA and (eluent B) 0.050% TFA in acetonitrile-water 80/20 (v/v). The applied gradient was linear from 0% to 55% B in 40 min, at a flow rate of 0.30 ml/min. The T splitter permitted 0.20 ml/min to flow toward the diode array detector and 0.10 ml/min toward the ESI source. The photodiode array detector was set at a wavelength of 214 and 276 nm. During the first 5 min of separation eluate was not addressed to the mass spectrometer to avoid source contamination and instrument damage because of the high salt concentration. Mass spectra were collected every 3 ms in the positive ion mode. MS spray voltage was 4.50 kV and capillary temperature was 220 °C.
High-resolution nano-HPLC-ESI-MS/MS experiments were performed by using the following eluents: (A) 0.05% (v/v) aqueous TFA and (B) 0.05% (v/v) TFA in acetonitrile. The applied gradient was 0–4 min 5% B, 4–34 min from 5% to 50% B (linear), 34–64 min from 50% to 90% B (linear), at a flow rate of 4.5 μl/min. High-resolution positive MS/MS spectra were collected in full scan using the lock mass for internal mass calibration (polydimethyl cyclosiloxane, 445.1200 m/z) with the resolution of 60,000 and 30,000, respectively, and m/z range from 350 to 2000. In data-dependent acquisition mode the three most intense multiply charged ions were selected and fragmented by using collision induced dissociation (35% normalized collision energy) and spectra were recorded. Alternatively, fragmentation was carried out under the same conditions on selected multiply charged ions corresponding to specific protein masses. Tuning parameters were: capillary temperature 220 °C, source voltage 2.4 kV, capillary voltage 26 V, tube lens voltage 245 V.
MALDI-TOF-MS Analysis
For MALDI-TOF-MS analysis, samples were dissolved in 0.1% aqueous TFA and the solution treated with a C-18 ZipTip pipette tip (Millipore, Billerica, MA) following the manufacturer indications including a wash with 0.1% TFA and elution with 0.1% TFA/acetonitrile (1:1, v/v). The desalted solution was mixed 1:1 (v/v) with saturated solutions of R-cyano-4-hydroxycinnamic acid in acetonitrile/water (50:50, v/v) containing 0.1% TFA. Aliquots of 1 μl of the mixture were spotted onto the stainless steel target of the MALDI instrument. The calibration was performed using peptide calibration standards (angiotensin I and II, substance P, and bombesin, m/z range 1000–3150 Da). Positive MALDI spectra were acquired in either linear or reflectron mode with a pulsed nitrogen laser (337 nm). In linear mode an acceleration voltage of 20 kV, a detector gain voltage of 1300 V, a pulsed ion extraction time of 350 ns and a laser frequency of 5 Hz were applied. In the reflectron mode an acceleration voltage of 19 kV, a detector gain voltage of 1400 V, a pulsed ion extraction time of 150 ns, and a laser frequency of 5 Hz were applied. Mass spectra were acquired over the mass range of 700–6000 Da with the low mass cut-off of 500 Da and 400 scans were averaged for each spectrum.
Preparative RP-HPLC Purification of the Proteins Detectable in Whole Saliva of Preterm Newborns
Semipreparative RP-HPLC was utilized in order to purify the proteins detected in whole saliva of preterm newborn and/or to collect peptide fragments from trypsin digest (see in the following sections). Acidic solution from preterm saliva (200 μl or more, when available) or tryptic digest were purified on a Surveyor Plus LC system (ThermoFisher) equipped with a PDA detector set at 214 and 280 nm. The column was a Vydac C8 with 5 μm particle diameter (column dimensions 250 × 4.6 mm). Eluent A was 0.056% aqueous TFA and eluent B was 0.050% TFA in acetonitrile-water 80/20 (v/v). The applied gradient was linear from 0% to 55% B in 40 min, at a flow rate of 0.90 ml/min. Fractions were collected in concomitance with the peak exit. The content of any purified fraction was checked by using HPLC-ESI-MS procedures described in the previous sections. Fractions with similar content obtained from different HPLC separation were sometimes pooled. Several fractions resulted to contain more than one protein. They were used for enzymatic and chemical treatments as described in the following sections without further purification.
Automated Edman Sequencing
Protein and peptide sequencing was carried out on the purified protein or peptide with a Procise 610A Protein Sequencer (Applied Biosystems, Foster City, CA). Sequencing was carried out by classical Edman degradation according to manufacturer's instructions.
Enzymatic Dephosphorylation
Freeze-dried powder of the purified protein or of protein mixtures (ca. 50 μg) was dissolved in 100 μl of 0.2 m Tris-HCl (pH 8.6) and 40 μl of calf intestinal alkaline phosphatase (1 EU/μl, Roche-Boehringer, Mannheim, Germany) were added. Incubation was carried out at 37 °C and after 40 min the solution was centrifuged at 8000 × g for 5 min, and the supernatant immediately analyzed by RP-HPLC-ESI-MS.
Removal of N-Terminal Acetylation
Freeze-dried powder of the purified protein or of protein mixtures was incubated in the presence of 25% TFA at 55 °C for 1h and the solution was analyzed by RP-HPLC ESI-MS.
Reduction and Alkylation of Cysteine Residues
Freeze-dried powders of the purified proteins or protein mixtures were dissolved at a concentration about 5 μmol/L in 100 μl of 100 mmol/L Tris-HCl buffer pH 7.5 containing 5 mmol/L dithiotreitol. The solution was incubated at 100 °C for 5 min and subsequently at 50 °C for 15 min. After incubation, 1 μl of dimethylsulfoxide and 13 μl of 0.4 mol/L 4-vinylpyridine in methanol 95% were added at room temperature to the solution. The solution was kept in the dark at room temperature for 3 h. Reaction was stopped by adding 115 μl of 0.2% TFA in H2O. The solution was analyzed by RP-HPLC ESI-MS.
Trypsin Digestion
50 μg of the freeze-dried powder of the purified protein or protein mixture was dissolved in 250 μl of 0.5 m ammonium hydrogen-carbonate (pH 8.0) and digested at 37 °C under stirring for 5 h by adding 10 units of immobilized trypsin (Pierce Biotechnology, Rockford, IL). After incubation the solution was centrifuged at 8000 × g for 5 min and the supernatant lyophilized and stored at −80 °C.
Data Analysis
Deconvolution of averaged ESI mass spectra was automatically performed either by the software provided with the Deca-XP instrument (Bioworks Browser) or by MagTran 1.0 software (11). Experimental mass values were compared with theoretical average mass (Mav) values available at the Swiss-Prot data bank (http://us.expasy.org/tools). Proteins and derivatives were identified as described in the result section.
The relative abundances of proteins and derivatives were determined by measuring the XIC peak area and relating it to 1.0 ml of whole saliva. Under identical experimental conditions this value is linearly proportional to peptide concentration and it can be used with confidence to monitor relative abundances (12). In the determination of XIC peak area a correct choice of the m/z values for protein detection is necessary, avoiding m/z potentially overlapping with ESI spectra of other close-eluting proteins in crowded chromatographic elution ranges (see “Results”). The window for all the m/z values chosen was in a range of ± 0.5 m/z. The percentage error of the measurements was less than 10%. Pearson r coefficient was used to evaluate linear correlations among the (XIC peak area/ml of whole saliva) of different proteins. Data obtained from the analysis of trypsin digestion products by nano-HPLC-ESI-MS/MS LTQ Orbitrap XL apparatus were elaborated by the Proteome Discoverer 1.0 program, based on SEQUEST cluster as search engine (University of Washington, USA, licensed to Thermo Electron Corp., San Jose, CA) against Swiss-Prot human proteome (March 10th, 2010 release; uniprot-taxonomy-9606-AND-reviewed-yes.fasta; 34756 nonredundant protein sequences). For peptide matching the following limits were used: Xcorr scores greater than 1.5 for singly charged peptide ions and 2.0 and 2.5 for doubly and triply charged ions, respectively, one missed cleavage sites. Different searches were carried out allowing the recognition of various post-translational modifications (PTMs). Precursor mass search tolerance was set to 10 ppm and fragment mass tolerance was set to 1.5 Da.
RESULTS
Top-Down HPLC-ESI-MS Analysis of Whole Saliva of Preterm Newborns
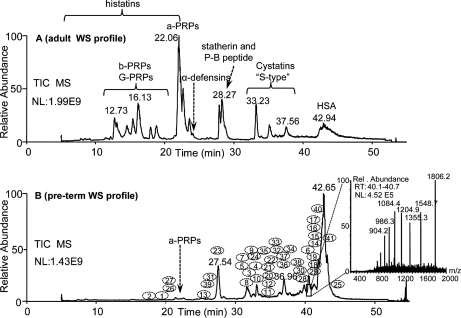
The acidic soluble fraction of whole saliva of preterm human newborns (195–220 days of postconceptional age) showed an HPLC-ESI-MS total ion current profile very different from that of the adult (Fig. 1, A and B). In the profile of saliva from adult (Fig. 1A) the principal chromatographic peaks corresponded to peptides and proteins specific of salivary glands (acidic and basic proline-rich proteins, statherin, histatins, and S-type cystatins) and to their fragments and derivatives (13–18). In the profile of saliva from preterm newborns (Fig. 1B) proteins characteristic of adult whole saliva were either absent or present in very low relative amounts. On the contrary, various proteins almost absent in adult whole saliva were consistently detected in whole saliva of preterm newborns, as evidenced by deconvolution of their ESI-MS spectra. For example, the indent of Fig. 1 shows the ESI mass spectrum of the protein, identified as S100A8, eluting between 40.0 and 40.8 min, which following deconvolution displayed an experimental average mass (Mav) value of 10834 ± 2 Da. The numbers assigned to specific elution positions in the B profile correspond to the protein masses listed in Tables I and II. The numbers 40 and 41 were assigned to α-globin (Swiss-Prot Number: P69905; Mav 15128 ± 3 Da) and γ-globins (P69891/2; Mav Aγ: 16009 ± 3 Da; Gγ: 15995 ± 3 Da) (Table III). Table I shows the 25 Mav values of the salivary proteins observed with high frequency (> 50%) in 61 specimens of whole saliva collected at different postconceptional age from eight preterm newborns. Those detected with low frequency (10%–50%) are reported in Table II. In Tables I and II protein elution times are also reported.
Fig. 1.
Comparison between the total ion current HPLC-ESI-MS profiles of the acidic soluble fraction of whole saliva from an adult (34 years, panel A) and a preterm newborn (226 days of postconceptional age, Panel B). In the adult profile, the elution ranges of the main families of secretory salivary proteins are reported (a-PRPs, b-PRPs, G-PRPs: acidic, basic, glycosylated proline-rich proteins. HSA: human serum albumin). In the preterm profile the numbers 1–41 show the chromatographic positions of the proteins, listed according to Tables I and II. Numbers 40 and 41 (not reported in Tables I and II) refer to α-globin and γ-globins, respectively. The indent in panel B shows, for example, the ESI spectrum of the protein eluting between 40.0 and 40.8 min, which after deconvolution displayed an experimental Mav of 10834 ± 2 Da and that was identified as S100A8. Whole saliva (WS), NL, normalization level; RT, retention time.
Table I. Proteins masses (Da) detected with high frequency (more than 50%) in the HPLC-ESI-MS profile of saliva from preterm newborns, and their attributions, Swiss-Prot Numbers, elution times and frequencies. Experimental error of the average mass: ± 0.02%.
| Mav exp. | Identity | Swiss-Prot number | Elution time (min) | Frequency (8 subjects) | Frequency (61 samples) | |
|---|---|---|---|---|---|---|
| 1 | 4936.5 | Thymosin β10 | P63313 | 19.9–21.7 | 8/8 | 60/61 |
| 2 | 4963.5 | Thymosin β4 | P62328 | 18.1–18.9 | 8/8 | 60/61 |
| 3 | 9956 | Non identified (n.i.) | 31.7–32.3 | 8/8 | 57/61 | |
| 4 | 10651 | n.i. | 32.9–33.5 | 8/8 | 44/61 | |
| 5 | 10765 | n.i. | 31.2–32.0 | 8/8 | 56/61 | |
| 6 | 10834 | S100A8a | P05109 | 40.0–40.8 | 8/8 | 61/61 |
| 7 | 10875 | n.i. | 32.2–32.8 | 8/8 | 34/61 | |
| 8 | 11006 | Stefin A | P01040 | 31.4–32.2 | 8/8 | 61/61 |
| 9 | 11049 | n.i. | 32.1–32.9 | 8/8 | 56/61 | |
| 10 | 11182 | Stefin B | P04080 | 32.6–33.4 | 8/8 | 48/61 |
| 11 | 11368 | S100A7a (D27 var.) | P31151 | 36.7–37.3 | 8/8 | 48/61 |
| 12 | 11382 | S100A7a (E27 var.) | P31151 | 36.6–37.2 | 8/8 | 44/61 |
| 13 | 11710 | Antileukoproteinase | P03973 | 25.8–26.6 | 8/8 | 55/61 |
| 14 | 12689 | S100A9a | P06702 | 41.8–42.6 | 8/8 | 59/61 |
| 15 | 12769 | S100A9a | P06702 | 41.8–42.6 | 8/8 | 59/61 |
| 16 | 13153 | S100A9a | P06702 | 41.5–42.3 | 8/8 | 44/61 |
| 17 | 13233 | S100A9a | P06702 | 41.5–42.3 | 8/8 | 42/61 |
| 18 | 13274 | n.i. | 41.2–42.0 | 8/8 | 48/61 | |
| 19 | 13456 | n.i. | 41.1–41.9 | 8/8 | 55/61 | |
| 20 | 14694 | Lysozyme | P61626 | 35.2–36.0 | 8/8 | 58/61 |
| 21 | 14990 | n.i. | 35.0–35.8 | 8/8 | 31/61 | |
| 22 | 15079 | n.i. | 34.2–35.0 | 8/8 | 43/61 | |
| 23 | 17239 | SPRR3 (isof. 17 kDa) | A5YKK8 | 27.0–27.8 | 8/8 (7/8) | 49/61 (41/61) |
| “ | 18065 | SPRR3 (isof. 18 kDa) | Q9UBC9 | 27.2–28.0 | (5/8) | (28/61) |
| 24 | 22365 | Histone H1c | P16402 | 34.0–34.5 | 8/8 | 52/61 |
| 25 | 24652 | n.i. | 44.4–45.2 | 8/8 | 46/61 |
a For the nomenclature of S100 proteins see Ref. 19.
Table II. Protein masses detected with low frequency (between 10–50%) in the HPLC-ESI-MS profile of saliva from preterm newborns, and elution times and frequencies. Experimental error of the average mass: ± 0.02%.
| Mav exp. | Identity | Swiss-Prot number | Elution time (min) | Frequency (8 subjects) | Frequency (61 samples) | |
|---|---|---|---|---|---|---|
| 26 | 7806 | n.i. | 20.2–21.7 | 7/8 | 17/61 | |
| 27 | 7867 | n.i. | 20.2–21.7 | 7/8 | 17/61 | |
| 28 | 10444 | S100A12 | P80511 | 39.8–40.2 | 8/8 | 20/61 |
| 29 | 11652 | S100A11 | P31949 | 42.4–43.1 | 4/8 | 15/61 |
| 30 | 13780 | n.i. | 39.0–39.7 | 6/8 | 24/61 | |
| 31 | 16001 | n.i. | 27.0–27.7 | 7/8 | 18/61 | |
| 32 | 17803 | n.i. | 36.0–36.8 | 5/8 | 8/61 | |
| 33 | 18420 | n.i. | 35.8–36.6 | 6/8 | 15/61 | |
| 34 | 20206 | n.i. | 37.6–38.1 | 4/8 | 8/61 | |
| 35 | 20930 | n.i. | 33.4–33.8 | 7/8 | 15/61 | |
| 36 | 22698 | n.i. | 36.5–37.3 | 8/8 | 17/61 | |
| 37 | 22776 | n.i. | 36.5–37.3 | 8/8 | 16/61 | |
| 38 | 24904 | Peroxiredoxin 6 (?) | P30041 | 39.6–40.4 | 6/8 | 14/61 |
| 39 | 27050 | n.i. | 26.4–27.0 | 3/8 | 7/61 |
Table III. Different experimental approaches carried out for the identification of the proteins listed in Table I and II and post-translational modifications characterized.
A, high resolution MS/MS of the intact protein; B, coincidence with standards; C, N-terminal Edman sequencing; D, peptide mass fingerprint (trypsin) by MALDI-TOF-MS and/or by ESI-MS; E, MS/MS of the trypsin digest; F, cysteine alkylation by vinyl-pyridine; G, removal of N-terminal acetyl group; H, phosphatase treatment.
| Identity | N-term loss | N-term acetyl. | Other | Exp appr. | Mav exp.a | Mav theor.b | Mav theor.c | |
|---|---|---|---|---|---|---|---|---|
| 1 | Thymosin β10 | M | Yes | A,B | 4936.5 | 4936.5 | 4936.5 | |
| 2 | Thymosin β4 | M | Yes | A,B | 4963.5 | 4963.5 | 4963.5 | |
| 6 | S100A8 | - | No | A | 10834 | 10834 | 10834 | |
| 8 | Stefin A | - | No | A,D | 11006 | 11006 | 10875 | |
| 10 | Stefin B | - | Yes | A | 11182 | 11182 | 11182 | |
| 11 | S100A7 (D27 var.) | M | Yes | A | 11368 | 11368 | 11326 | |
| 12 | S100A7 (E27 var.) | M | Yes | A | 11382 | 11382 | 11382 | |
| 13 | Antileukoproteinase | - | No | 8 S-S | A,C | 11710 | 11710 | 11710 |
| 14 | S100A9 | MTCKM | Yes | A,D,E | 12689 | 12689 | - | |
| 15 | S100A9 | MTCKM | Yes | Phos | G,H | 12769 | 12769 | - |
| 16 | S100A9 | M | Yes | A,D,E | 13153 | 13153 | 13111 | |
| 17 | S100A9 | M | Yes | Phos | G,H | 13233 | 13233 | 13191 |
| 20 | Lysozyme | - | No | 4 S-S | C,F | 14694 | 14694 | 14694 |
| 23 | SPRR3 (17 kDa) | M | Yes | 7 cys | D,E,F,G | 17239 | 17239 | 17328 |
| SPRR3 (18 kDa) | M | Yes | 8 cys | D,E | 18065 | 18065 | 18154 | |
| 24 | Histone H1c | ? | ? | ? | D | 22365 | ? | 22260 |
| 28 | S100A12 | M | No | A | 10444 | 10444 | 10444 | |
| 29 | S100A11 | M | Yes | A | 11651 | 11651 | 11740 | |
| 38 | Peroxiredoxin 6 (?) | - | - | - | 24904 | 24904 | 24904 | |
| 40 | α-globin | M | - | B,D,E | 15128 | 15128 | 15128 | |
| 41 | γ-globinsd | M | - | B,D,E | 16009 | 16009 | 16009 |
a Experimental error of the average mass: ± 0.02%.
b Theoretical average mass computed considering the PTMs reported in the table.
c Theoretical average mass available in Swiss-Prot data-bank (in italics when the theoretical Mav reported in Swiss-Prot did not correspond to the experimental).
d Only the average mass of Aγ globin is reported. The Mav of Gγ globin is 15995 Da.
The detection of γ-globins required sometimes the modification of the elution gradient because of overlapping with heterogeneous glyco-proteins, which generated crowded ESI spectra.
Identification of the Proteins Detected in Saliva of Preterm Newborns
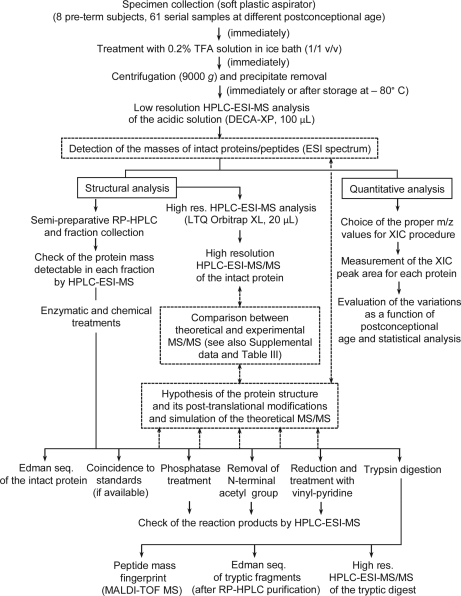
Because of the small sample volume available, the most demanding task of this study was the characterization of the proteins reported in Tables I and II. The Mav value of the intact protein, even if determined with good accuracy (in our apparatus ± 0.02%), is obviously not enough for the definitive identification. The experimental scheme of the top-down proteomic approach followed in this study is described in the flow chart of Fig. 2. Preparative RP-HPLC on some samples of preterm newborn saliva allowed several chromatographic fractions to be collected. Fractions were analyzed by HPLC-ESI-MS in order to check protein content, and, because of similar polarity, more than one protein/peptide in each chromatographic fraction was often detected. Fractions showing a predominant protein were lyophilized and an aliquot subjected to automated Edman sequencing. It was possible to sequence only lysozyme and antileukoproteinase whereas results of the other proteins suggested a blocked N-terminal residue. Some of the chromatographic fractions were subjected to reduction and alkylation with vinyl-pyridine, others were treated to remove the N-terminal acetyl group, others were submitted to the action of phosphatases, following the procedures described in the Experimental Procedures section. After these reactions, an aliquot of the modified proteins was analyzed by HPLC-ESI-MS in order to measure the mass variation. Another approach for protein characterization was based on trypsin digestion of purified proteins and analysis of digestion products by nano-HPLC-ESI-MS/MS using the high-resolution LTQ Orbitrap XL apparatus. Many MS/MS fragmentation spectra were manually analyzed. This approach sometimes resulted in plausible protein identification. However, often theoretical Mav values deduced from the sequence reported in the data bank did not correspond to the experimental one. For some proteins the hypothesis of loss of the N-terminal methionine residue followed by N-terminal acetylation resulted in a good correspondence of the theoretical Mav with the experimental one. Protein structures were definitely confirmed by submitting intact protein to high-resolution nano-HPLC-ESI-MS/MS experiments (Fig. 2), performed on the most abundant ions. Sometimes this procedure provided an MS/MS fragmentation spectrum rich enough to be manually compared with the theoretical spectrum generated by the MS-Product program available at the Protein-Prospector site (http://prospector.ucsf.edu/prospector/mshome.htm). Structures were assumed as correct if the experimental fragments with a relative abundance higher than 10% were present in the theoretical fragmentation spectrum with 0.03 Da fragment mass tolerance (see Supplemental Data).
Fig. 2.
Flow chart of the top-down strategy applied in this study. The experimental flow is described by solid arrows, lines, and boxes. Steps in dashed boxes connected by dashed arrows allow the unambiguous protein identification, which requires the correspondence of experimental Mav with that one of the structure hypothesized and the coincidence between experimental and theoretical MS/MS spectra (within the experimental error). Detailed information on each characterized protein is reported in Table III and in Supplemental data.
Table III lists the experimental and theoretical mass values, the number of accessible cysteins, the proposed PTMs, and the methodologies utilized to establish protein identity. Proteins with Mav values corresponding to the values available in Swiss-Prot data bank were Tβ4 and Tβ10, S100A8 (calgranulin A; for S100 nomenclature see reference (19)), stefin B, lysozyme C, S100A12 (calgranulin C), antileukoproteinase, peroxyredoxin 6 (this last pending for a definitive characterization), α-globin and γ-globins. The protein with a Mav 11,006 ± 2 Da was identified as stefin A with an additional N-terminal methionine residue (see Supplemental Data) with respect to the isoform reported in Swiss-Prot (Mav 10,875 Da). We detected just following the stefin A peak (Table I) a protein with an experimental mass of 10,875 ± 2 Da but the MS/MS fragmentation spectrum of the intact protein, even poor, did not correspond to the theoretical fragmentation values expected for stefin A missing N-terminal methionine. S100A7 (psoriasin) was detected in two isoforms, one (Mav 11,382 ± 2 Da) corresponding to the S100A7 variant reported in Swiss Prot data bank. The other (Mav 11,368 ± 2 Da), which was the most abundant, corresponded to the variant E27→D. Both S100A7 variants were found to be submitted to N-terminal acetylation following the loss of the terminal methionine. The Mav values of the other proteins reported in Table III were in agreement with different PTMs not reported in the data banks. S100A9 (calgranulin B) was detected in four isoforms already characterized by other authors in human granulocytes (20). Two isoforms (Mav 13,153 ± 2 and 13,233 ± 2 Da), which could be defined as long-types, were found to be acetylated following loss of the N-terminal methionine residue and differed from each other in phosphorylation of the penultimate threonine residue of the sequence (Thr112). The other two isoforms (Mav 12,689 ± 2 and 12,769 ± 2 Da), defined as short-types, were found to be acetylated following the loss of the five N-terminal amino acid residues (MTCKM) and differed in the phosphorylation of the same residue of the long-types (Thr108). S100A11 (calgizzarin) was also found to be acetylated at the N-terminal residue following methionine loss (PTM not available in Swiss-Prot data bank). In a previous study (10) we were able to establish that the same PTMs (methionine loss and N-terminal acetylation) were responsible for the observed Mav values of the two isoforms of SPRR3. The isoform with Mav 18,065 ± 3 Da differed for the insertion of a further octapeptide repeat (GCTKVPEP) to the five identical repeats present in the 17,239 ± 3 Da isoform and for the presence of Leu at positions 148, instead of Val at the corresponding position 140 of the shorter isoform (10). Three subjects (out of 8) were homozygous for the 17,239 ± 3 Da isoform, one was homozygous for the 18,065 ± 3 Da isoform, and four subjects were heterozygous. No polymorphism was observed for the other salivary components of preterm newborns (except for γ globin, which was detected, obviously, as Aγ and Gγ isoforms). Tentative identification of the protein with Mav 22,365 ± 4 Da with histone H1c (histone H1.3) was based on analysis of the tryptic digestion mixture obtained from the purified protein. Data analysis by Proteome Discoverer attributed three peptides to histone H1c, and this assignment was also manually checked. The three fragments corresponded to the sequences 34–46, 65–75, and 140–148 of the protein, two present in other histones, and one (fr. 140–148) specific for histone H1c. However, the theoretical Mav of histone H1c is 22,219 Da, 146 ± 4 Da less than the experimental value. Different isoforms of H1 histone have been detected in the spleen of adult subjects (21). Thus, we may hypothesize either that the preterm newborn protein could be an unknown variant of histone H1c or that not characterized PTMs are present in the mature protein. Tentative identification of peroxiredoxin 6 was only based on the detection in saliva of preterm infants of a peptide corresponding to the fragment 1–32 of the intact protein. Therefore, this protein is still pending for a definitive identification. Detailed information on the results of the experiments performed to characterize proteins and PTMs reported in Table III is available in Supplemental Data.
Some other protein masses (not reported) were sporadically detected in whole saliva of preterm newborns and some masses, probably pertaining to peptide fragments of larger proteins, were also detected. Moreover, the (small) precipitate formed during the acidic treatment of whole saliva might contain some other proteins/peptides not detectable in the adult. Their characterization will require further studies.
Determination of Protein Relative Amount as a Function of Preterm Postconceptional Age
To study the time course of the identified proteins, HPLC-ESI-MS analyses of samples collected at different postconceptional age (from 194 to 545 days) from eight newborns, for a total of 61 samples, were carried out. The relative amounts of each protein were established, as previously reported (6), by using the area of the eXtracted Ion Current (XIC) peaks and relating it to 1 ml of whole saliva. Quantitative analysis is reported only for the 25 proteins with higher frequencies in the 61 samples listed in Table I. The multiply charged ions selected for XIC search of each protein are reported in Table IV.
Table IV. Charge of the ions utilized for the determination of the XIC peak area of the 25 proteins of Table I and parameters expressing concentration trend over time of each protein in the population investigated. Experimental error of the average mass: ± 0.02%. Ratios between the mean of (XIC peak area)/(ml of whole saliva) values (reported in Fig. 3) measured in three different postconceptional age (PCA) ranges: 194–235, 236–290, and 291–545 days of postconceptional age.
| Mav exp. | Identity | Charge of ions | Apex (PCA) | (m 236–290)/(m 194–235) | (m 291–545)/(m 236–290) | |
|---|---|---|---|---|---|---|
| 1 | 4936.5 | Thymosin β10 | From 3+ to 5+ | <195 | 0.46 | 0.15 |
| 2 | 4963.5 | Thymosin β4 | From 3+ to 5+ | 140–160a | 0.48 | 0.12 |
| 3 | 9956 | n.i. | From 6+ to 9+ | <195–210 | 0.46 | 0.12 |
| 4 | 10651 | n.i. | From 8+ to 11+ | <195 | 0.25 | 0.05 |
| 5 | 10765 | n.i. | From 8+ to 11+ | <195 | 0.39 | 0.22 |
| 6 | 10834 | S100A8 | From 8+ to 11+ | <195 | 0.73 | 0.12 |
| 7 | 10872 | n.i. | From 8+ to 11+ | <195–210 | 0.16 | 0.02 |
| 8 | 11006 | Stefin A | From 8+ to 11+ | <195–210 | 0.45 | 0.25 |
| 9 | 11049 | n.i. | From 8+ to 11+ | <195 | 0.44 | 0.00 |
| 10 | 11182 | Stefin B | From 7+ to 10+ | <195–210 | 0.32 | 0.03 |
| 11 | 11368 | S100A7 (D27) | From 8+ to 11+ | 210–240 | 0.89 | 0.11 |
| 12 | 11382 | S100A7 (E27) | From 7+ to 10+ | 210–240 | 1.04 | 0.05 |
| 13 | 11710 | Antileukoproteinase | From 6+ to 9+ | 205–220 | 0.54 | 0.31 |
| 14 | 12690 | S100A9 short | From 9+ to 13+ | <195–220 | 0.58 | 0.13 |
| 15 | 12770 | S100A9 short (phos) | From 9+ to 13+ | <195–220 | 0.63 | 0.18 |
| 16 | 13155 | S100A9 long | From 10+ to 14+ | <195–205 | 0.27 | 0.05 |
| 17 | 13235 | S100A9 long (phos) | From 10+ to 14+ | <195–205 | 0.27 | 0.09 |
| 18 | 13274 | n.i. | From 10+ to 14+ | <195–220 | 0.65 | 0.15 |
| 19 | 13461 | n.i. | From 10+ to 14+ | 210–240 | 0.94 | 0.13 |
| 20 | 14694 | Lysozyme | From 8+ to 12+ | 210–240 | 1.00 | 0.40 |
| 21 | 14990 | n.i. | From 12+ to 16+ | <195 | 0.41 | 0.06 |
| 22 | 15079 | n.i. | From 12+ to 16+ | <195–220 | 0.45 | 0.02 |
| 23 | 17239 | SPRR3 | From 12+ to 17+ | <195 | 0.19 | 0.09 |
| 18065 | ||||||
| 24 | 22365 | Histone H1c | From 16+ to 22+ | <195–220 | 0.72 | 0.22 |
| 25 | 24652 | n.i. | From 14+ to 20+ | 230–260 | 2.50 | 0.04 |
a Value from Ref. (8).
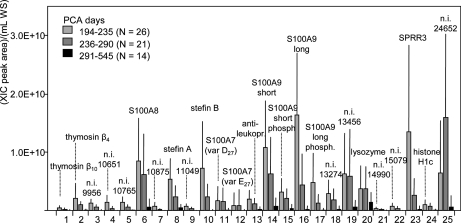
Fig. 3 shows the mean values of the ratio (XIC peak area)/(ml of whole saliva (WS)) computed in three ranges of postconceptional age, i.e. 194–235 days (n = 26), 236–290 days (n = 21), and 291–545 days (n = 14) for all the 25 proteins. The values reported for SPRR3 correspond to the sum of the two isoforms. In the heterozygous, the XIC peak areas of the two isoforms were always very similar.
Fig. 3.
Mean values of the ratios (XIC peak area)/(ml of WS) calculated for the 25 proteins detected in saliva of preterm newborns. Mean values refer to three groups of saliva samples subdivided according to postconceptional age of preterm newborns: 194–235 (n = 26), 236–290 (n = 21), 291–545 days (n = 14). Bars correspond to standard deviation values. SPRR3 data refer to both isoforms. WS: whole saliva.
Even though the values reported in Fig. 3 do not correspond to absolute concentration values, because ESI efficiency differs between proteins, they suggest that concentrations of the proteins were markedly different. In previous studies, we correlated the values of the XIC peak area of Tβ4 and Tβ10 to their concentration (8, 9). The mean concentration of Tβ4 in saliva of preterm newborns with 194–235 days of postconceptional age was about 1 μmol/L, and that one of Tβ10 was about four times lower. Because of the high structural similarity of isoforms and variants, data reported in Fig. 3 suggest that concentration of the D27 variant of S100A7 is at least three times higher than the E27 variant.
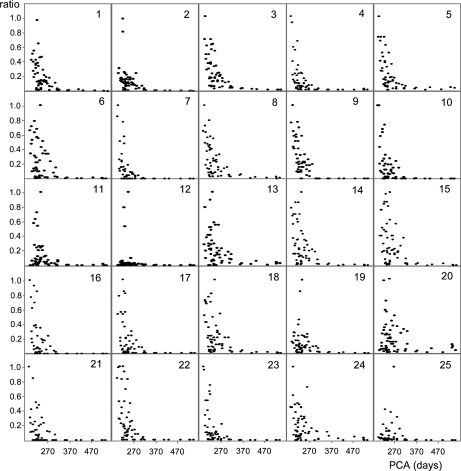
In order to compare the concentration variation over time of the various proteins reported in Table I, quantitative data obtained in the 61 samples were normalized to the highest value determined for each protein (Fig. 4). Fig. 4 shows that concentration of the majority of the proteins decreased, even though with different slope. For instance, although stefin A (panel 8) concentration showed a fast decrease from 195 days of postconceptional age, S100A8 (panel 6) decrease was slower. The unidentified protein with Mav 24,652 ± 5 Da (panel 25) and S100A7 (D27) (panel 11) showed low concentration values immediately following birth followed by a fast increase within 1 or 2 weeks up to maximum of concentration. Thereafter, concentration decreased and reached the minimal levels around 260–280 days of postconceptional age (corresponding to the normal term of delivery). Panel 12, which shows data of S100A7 (E27), is compressed because of three high values detected in one subject. However, the shape is similar to that one of S100A7 (D27) isoform, but with values significantly lower. Further evidence from Fig. 4 is that concentration of all the proteins reached the minimal levels in correspondence with the normal term of delivery, sometimes disappearing, at the sensitivity level of the MS apparatus.
Fig. 4.
Ratios (XIC peak area)/(ml of WS) of the 25 proteins listed in Table I normalized to highest value determined for each protein in the 61 samples plotted as a function of postconceptional age. SPRR3 data refer to both isoforms. PCA, postconceptional age.
Fig. 4, showing the values measured in the 61 samples, does not allow the striking differences existing among subjects to be appreciated. Differences are evident from the large bars of the standard deviations of Fig. 3. The high interindividual variability becomes clear from Fig. 5, where the values of six selected proteins measured in three subjects are reported up to 290 days of postconceptional age. Ratios of (XIC peak area)/(ml of WS) reported in Fig. 5 were normalized with respect to the highest value determined in each subject in the temporal range considered, and therefore they are different from those reported in Fig. 4. Fig. 5 clearly shows different trends of decrease, some almost linear (i.e. Tβ4), others with exponential-like shapes, but with different slopes (i.e. stefin B and the not identified protein with a Mav of 10,651 ± 2 Da), others characterized by a sharp decrease following a constant apex period (i.e. short isoform of S100A9 and SPRR3), and others, such as S100A7 (D27), by a biphasic trend. Interestingly, Fig. 5 shows that the decrease of concentration of each protein showed similar shapes in the different subjects, but it was often phase shifted.
Fig. 5.
The ratios (XIC peak area)/(ml of WS) of six proteins from three preterm newborns (1–3) are reported as an example of interindividual variability. Ratios are normalized to highest value determined for each protein in each subject in the range of 194–290 days of postconceptional age. SPRR3 data refer to the 17 kDa isoform. PCA, postconceptional age.
Because of high interindividual variability temporal changes of the different proteins varied markedly in the 8 subjects. Table IV shows the postconceptional age ranges corresponding to the maximum concentration observed for each protein. When the level measured at the lowest postconceptional age was the highest, it was assumed that maximum concentration occurred before 195 days of postconceptional age. In order to find a parameter that could give for each protein a rough estimate of the concentration trend over time in the population investigated, we calculated the ratio between the mean concentration values shown in Fig. 3. The results reported in Table IV suggest in some cases time-coordinate functions, such as for stefin A and B whose decrease was similar, in other hierarchical functions, such as for S100A7, whose increase followed that of many other proteins.
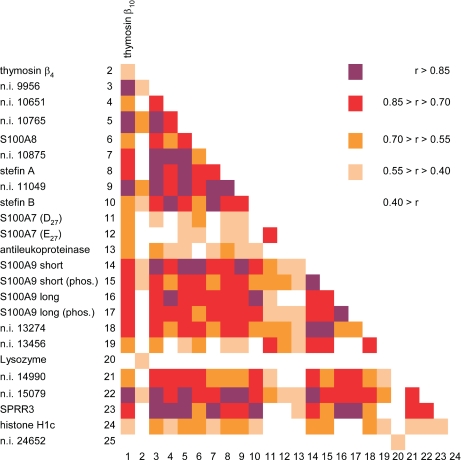
Fig. 6 shows the correlation coefficients (Pearson r) computed among all the proteins of Table I. Because of the size of our sample (n = 61), r values greater than 0.400 indicate a level of significance better than p < 0.001. Fig. 6 shows several clusters of high significant correlations, including the majority of S100 proteins, stefins, SPRR3, and some unidentified proteins. For its particular biphasic trend, S100A7 showed less significant correlations with the other proteins. Antileukoproteinase, lysozyme, and histone H1c showed low significant correlations. The unidentified protein with Mav 24,652 ± 5 Da showed a significant correlation only with lysozyme. Interestingly, although Tβ10 correlation with many proteins of the list was highly significant, the same was not observed for Tβ4. Interestingly, the two peptides showed not very high significant correlation each other. The highest correlation values (higher than 0.93) were observed between the two S100A9 isoforms and their phosphorylated counterparts, between S100A8 and S100A9 short type, as well as between SPRR3, stefin A, stefin B and the unidentified protein with Mav 10,651 ± 2 Da.
Fig. 6.
Colored map of the crossed correlation coefficients (Pearson r) computed among the 25 proteins of Table I. The significance at the conventional levels for r values greater than 0.400 is better than p < 0.001 (n = 61). The range of r values corresponding to the different colors is reported on the right side of the figure. SPRR3 data refer to both isoforms.
Extensive quantitative analysis of the proteins reported in Table II was not carried out, principally because of their low frequency. They were usually detectable in the samples in the range of 194–240 days of postconceptional age, disappearing in the total ion current profile at higher postconceptional age. This observation suggests that numerous other different proteins are probably present in fetal mouth during a temporal period preceding the one investigated in this study, with a maximum of expression occurring before 6 to-7 months of postconceptional age.
Comparison of the Salivary Protein Patterns of At-Term Newborns with Adults
As shown in Fig. 4, concentration of all the proteins detected in this study decreases in the range 195–280 days of postconceptional age reaching a value up to two orders of magnitude lower than the maximum measured. For all the proteins reported in Tables I and II the mean values measured in the range 270–280 days of postconceptional age (considered as the normal delivery time) corresponded perfectly to the mean values measured in whole saliva of at term newborns (n = 4). The mean values measured following 350 days of postconceptional age were very similar to those measured in adult whole saliva (n = 20). Often the protein was not detectable at all in adult whole saliva, even by XIC search, at the sensitivity level of our HPLC-MS apparatus. Statistical analysis of all the data collected did not disclose gender related differences.
DISCUSSION
The top-down proteomic approach used in this study gave relevant qualitative and quantitative information on the salivary proteome of human preterm newborns. In contrast to bottom-up proteomic strategies, knowledge of the mass of the intact protein obtained by MS measurements allows to discriminate between different isoforms. Clearly, the top-down strategy used in this study appears the most direct approach to detect the differences, e.g. between the isoforms of S100A7 and S100A9 proteins.
It should be mentioned that the proteins characterized in this study are likely to exert their function throughout a subtle interplay between monomeric and homo- and hetero-oligomeric not-covalent assemblies, which are unstable under the acidic conditions used during HPLC-ESI-MS analysis. Moreover, the acidic treatment generates a small precipitate, which in adult whole saliva is mainly constituted by mucins. In the preterm whole saliva, the precipitate can enclose other proteins relevant for the fetal development, which will be the aim of future studies.
Saliva is a body fluid of a very complex composition with fundamental roles in the protection of the mouth and its annexes during life. Although the precise molecular interplay between the different salivary adult components is far from being elucidated, the exploitation of this function is partly granted by specific peptides and proteins secreted by salivary glands that could play relevant roles in the protection of the enamel surface, in the assumption of food and beverage and in the modulation of the oral flora (1). This protection is not necessary during fetal life because fetus nutrients are supplied directly by the placental system of the mother and therefore protected against the threats of external environment. Until now the fetal mouth has been believed to be a passive organ reaching the physiological-anatomical maturation mostly following delivery. However, our group recently reported high levels of Tβ4 and Tβ10 in preterm newborn saliva (8). We were able to show that these peptides in fetuses mainly originate from salivary gland secretion, in contrast to adult saliva where low quantities originate from the gingival crevicular fluid (9). Thus, salivary glands may have a specific function during the intrauterine life. This view is further supported by this study, where we could show that several other proteins were present in whole saliva of preterm newborns. Salivary concentration of these proteins decreased as a function of postconceptional age, reaching values observed in at term newborns at about 270 days of postconceptional age and the values observed in adult whole saliva later in development.
The decrease of concentration as a function of preterm newborn age can be partly linked to an increased salivary flow-rate, which could not be measured for ethical reasons. However, because the shape of decrease for many proteins detected in this study was different, the variations are probably not linked to a dilution process but rather suggest a coordinate and hierarchical actions of these proteins. The interindividual variability shown in Fig. 5 most likely is not linked to feeding habits because of similar nutritional program, standardized for all the subjects.
Among the proteins characterized, many are members of the S100 protein family, which is the largest subgroup within the superfamily of EF-hand calcium-binding proteins, exclusively expressed in vertebrate (22, 23). A specific feature of this protein family is that some members are secreted from cells upon stimulation exerting cytokine- and chemokine-like extracellular activities via the receptor for advanced glycation endproducts. Functional diversification of this family of proteins is achieved by specific cell- and tissue-expression patterns, structural variations, different metal-ion binding properties, as well as by their ability to form homo- and hetero- and even oligomeric assemblies (24). In this study significant amounts of S100A7, A8, A9, A11, and A12 were detected in whole saliva of preterm newborns. Interestingly, the most abundant S100A7 isoform detected was the D27, with minor quantities of the E27 isoform. Some of the mass values not identified could pertain to other S100, such as the protein with a Mav 10,651 ± 2 Da, which might correspond to S100B or S100A5. Minute amounts of a protein with a mass of 11,712 ± 2 Da, which might correspond to S100A16, eluting in the proximity of S100A7, were found in few samples.
The presence of high amounts of two isoforms of SPRR3 in preterm whole saliva is intriguing, and it may be in some way related to the S100 family. The molecular mechanisms underlying the formation of the epithelial barrier start from the desmosomal cadherins, that are linked to the keratin cytoskeleton via several plaque proteins, such as desmoplakin and γ-catenin. However, oral keratinocytes express additional differentiation markers, including filaggrinin and tricohyalin, which associate with the cytoskeleton during terminal differentiation. Moreover, other proteins such as loricrin, involucrin, and small proline-rich proteins, comprising SPRR3, are cross-linked into the epithelial envelope by transglutaminases (25). The presence of SPRR3 in preterm whole saliva could be thus linked to the inability of immature keratinocytes to cross-link the protein to the cytoskeleton for a lack of epithelial transglutaminase. However, in this study no other proteins of the cytoskeleton were detected in whole saliva of preterm newborns and none of the protein masses waiting for characterization seems to correspond to other proteins of the cytoskeleton. Free SPRR3 is only detected in highly differentiated keratinocytes of well-differentiated squamous cell carcinomas, indicating that the protein is induced in skin tumors only when keratinocytes undergo extensive squamous differentiation (26). Furthermore, various authors have suggested that the proteins of the SPRR3 family (SPR family, pancornulins) play a relevant “active” role in the formation of cornified epithelia (27). The correlation between SPRR3, and the proteins of S100 family is interesting, considering that several of these proteins are related (have an EF-hand structural motif) and their genes localized in a cluster on human chromosome 1q21, which encodes for this protein (SPRR3) as well as for S100A7, A8, A9, A10, A11, and loricrin (19, 28, 29). Because of the presence of different transglutaminase reactive residues, S100A10 and S100A11 are incorporated into the cornified envelope of keratinocytes (30, 31). The colocalization of genes expressed during maturation of epidermal cells together with genes encoding calcium-binding proteins is particularly intriguing, because calcium levels tightly control the differentiation of epithelial cells, the expression of genes encoding epidermal structural proteins, as well as transglutaminase activity. Because of this close functional cooperation, the cluster 1q21, including these loci, has been proposed to be a specific gene complex and has been named “epidermal differentiation complex” (32).
The detection of stefin A and stefin B in whole saliva of preterm newborns can be related to some recent hypothesized properties of these proteins. Stefins A and B (or cystatins A and B) are indeed the better known members of type 1 cystatin family (cysteine proteinase inhibitors), generically called stefins (33). Stefins were commonly considered potent intracellular inhibitors of papain and cathepsins L, S, and H, but they have also been detected in significant amounts in different body fluids (33, 34) and recent studies outlined a very complex set of functions and interplays for them (35). Stefin A is predominantly present in blood and epithelial cells, particularly in high amounts in immune follicular dendritic cells, where it has been speculated that it plays a role in the prevention of apoptosis of B-lymphocytes by inhibition of cathepsin B (36, 37). A recent proteomic study postulated relevant functions of stefin A in mouse neonatal skin during development and in immune response (38), role probably related to the significant presence of stefin A found in this study in preterm whole saliva. Some authors have established the participation of this protein in the structural organization of cornified cell envelope too (39).
Stefin B is more widely distributed inside the cytoplasm of most human cells and increased levels have been described in a variety of malignant tumors (40). Its gene expression is increased in human monocytes following stimulation by lipopolysaccharides (41) and it plays important and composite, although not defined, roles in neural stem cells and in differentiated neurons and astrocytes (42). Moreover, it is an intracellular modulator of rat bone reabsorption (43). However, a possible correlation between stefin A and B with other proteins such as SPRR3 and S100 proteins are presently not known.
Antileukoproteinase could also be involved in the development of mucosal epithelia. This protein is an inhibitor of granulocytic serine proteases, modulating granulocyte-endothelium interaction (44) and it seems involved in the formation of cartilage, because of its accumulation in normal but not in arthritic cartilage (45).
Tβ4 and Tβ10 are new entries in the repertoire of adult salivary proteins and peptides (9, 46). Beta thymosins are ubiquitous peptides with intra- and extracellular functions (47, 48), although the secretion pathway of these peptides is not fully understood (49). Tβ4 was shown to modify the rate of attachment and spreading of endothelial cells on matrix components inducing matrix metalloproteinases (50) and to stimulate the migration of human umbilical vein endothelial cells (51). Although Tβ4 is a potent enhancer of angiogenesis, Tβ10 inhibits it and changes of the two peptides concentration ratio can exert either positive or negative control (52, 53). As demonstrated by Reti and coworkers (54), Tβ4 plays an important role in suppressing the production of interleukin-8 following stimulation by tumor-necrosis factor α and it acts on the whole as antimicrobial, anti-inflammatory and anti-apoptotic peptide on gingival fibroblasts. The properties of Tβ10 are less known when compared with Tβ4. The results of this study confirm that the two peptides are present in significant amounts in preterm whole saliva, and at variance in adult saliva, they derive mainly from salivary gland secretion (8). Tβ4 andTβ10 could play a role in angiogenesis, which has also reported for some S100 proteins, mainly S100A4 (for recent review see (55)). The different biological role of these peptides may be at the basis of the low correlations observed among the levels of thymosins β and the other proteins (Fig. 6).
The presence of large amounts of histone H1c in preterm whole saliva is puzzling. Indeed, in the hypothesis of its release from cell shedding, other histones should have been present in saliva. Similar consideration can be made for lysozyme, because its defensive role against pathogens does not fit with its decrease observed as a function of postconceptional age. The detection of α- and γ-globins was restricted to samples of all the subjects in the approximate range of 194–240 days of postconceptional age. Their detection most probably is because of their release from the preterm mucosal epithelium.
Many of the proteins identified in this study are considered to be tumor markers in the adult. This observation led us to suppose that during fetal development the interplay between these proteins might contribute to the molecular events responsible for cell growth and death. On the contrary, the abnormal regulation of their expression in the adult might be at the basis of anomalous cellular growth and might be connected to the development of different tumors with embryonal etiology. Moreover, the recognition of tumor stem cells in many solid cancers has reinvigorated the hypothesis of a pluripotent stem cell as the cell of origin for cancer (56). Because these tumor stem cells have direct access to embryologic programs, including the capacity to produce proteins and peptides normally secreted only during intrauterine life (57), we can speculate that at least a part of the fetal proteins described in this study, following disappearing in the postnatal life, could reappear in cancer cells. Preliminary data from our group on Tβ4 expression in salivary glands' tumors and in colon cancer (58) evidenced Tβ4 reactivity in tumor cells undergoing epithelial-mesenchymal transition, a highly conserved cellular program typical of several stages of embryonic development as well as of cancer invasion and metastasis (59). This finding seems to further support our hypothesis.
In conclusion, by analyzing a noninvasive specimen collection of saliva, this study suggests that the development of preterm newborns, and fetuses, requires the presence of distinct proteins of variable amounts at defined stages. Some of them have been characterized in this study, but others are waiting for a definitive identification. Further studies will be conducted to investigate their functions and interplay and to establish if they derive from gland secretion (as shown for Tβ4), from the fetal oral epithelium or other sources.
Acknowledgments
We acknowledge the financial support of Università di Cagliari, Università Cattolica in Rome, International Scientific Institute “Paolo VI” (ISI), MIUR, Italian National Research Council (CNR), Regione Sardegna, thanks to their programs of scientific research promotion and diffusion.
Footnotes
1 The abbreviations used are:
- SPRR3
- small proline-rich protein 3
- Mav
- average mass
- PTM
- post-translational modification
- Tβ4
- thymosin β4
- Tβ10
- thymosin β10
- XIC
- extracted ion current
- HPLC
- high pressure liquid chromatography
- RP
- reverse phase
- MS/MS
- tandem MS
- WS
- whole saliva
- HPLC-ESI-MS
- high performance liquid chromatography-electrospray ionization-mass spectrometry.
REFERENCES
- 1. Amerongen A. V., Veerman E. C. (2002) Saliva: the defender of the oral cavity. Oral Dis. 8, 12–22 [DOI] [PubMed] [Google Scholar]
- 2. Helmerhorst E. J., Oppenheim F. G. (2007) Saliva: a dynamic proteome. J. Dent. Res. 86, 680–693 [DOI] [PubMed] [Google Scholar]
- 3. Messana I., Inzitari R., Fanali C., Cabras T., Castagnola M. (2008) Facts and artifacts in proteomics of body fluids. What proteomics of saliva is telling us? J. Sep. Sci. 31, 1948–1963, Review [DOI] [PubMed] [Google Scholar]
- 4. Oppenheim F. G., Salih E., Siqueira W. L., Zhang W., Helmerhorst E. J. (2007) Salivary proteome and its genetic polymorphisms. Ann. N.Y. Acad. Sci. 1098, 22–50 [DOI] [PubMed] [Google Scholar]
- 5. Schipper R. G., Silletti E., Vingerhoeds M. H. (2007) Saliva as research material: biochemical, physicochemical and practical aspects. Arch. Oral. Biol. 52, 1114–1135 [DOI] [PubMed] [Google Scholar]
- 6. Cabras T., Pisano E., Boi R., Olianas A., Manconi B., Inzitari R., Fanali C., Giardina B., Castagnola M., Messana I. (2009) Age-dependent modifications of the human salivary secretory complex. J. Proteome Res. 8, 4126–4134 [DOI] [PubMed] [Google Scholar]
- 7. Inzitari R., Vento G., Capoluongo E., Boccacci S., Fanali C., Cabras T., Romagnoli C., Giardina B., Messana I., Castagnola M. (2007) Proteomic analysis of salivary acidic proline-rich proteins in human preterm and at term newborns. J. Proteome Res. 6, 1371–1377 [DOI] [PubMed] [Google Scholar]
- 8. Nemolato S., Messana I., Cabras T., Manconi B., Inzitari R., Fanali C., Vento G., Tirone C., Romagnoli C., Riva A., Fanni D., Di Felice E., Faa G., Castagnola M. (2009) Thymosin β4 and β10 levels in preterm newborn oral cavity and foetal salivary glands evidence a switch of secretion during foetal development. PLoS ONE. 4, e5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inzitari R., Cabras T., Pisano E., Fanali C., Manconi B., Scarano E., Fiorita A., Paludetti G., Manni A., Nemolato S., Faa G., Castagnola M., Messana I. (2009) HPLC-ESI-MS analysis of oral human fluids reveals that gingival crevicular fluid is the main source of thymosins beta4 and beta10. J. Sep. Sci. 32, 57–63 [DOI] [PubMed] [Google Scholar]
- 10. Manconi B., Cabras T., Pisano E., Nemolato S., Inzitari R., Iavarone F., Fanali C., Sanna M. T., Tirone C., Vento G., Romagnoli C., Faa G., Castagnola M., Messana I. (2010) Characterization of two isoforms of human SPRR3 from saliva of preterm human newborn and autoptic fetal oral mucosa, parotid and submandibular gland samples. Biochem. Biophys. Res. Comm. 398, 477–481 [DOI] [PubMed] [Google Scholar]
- 11. Zhang Z., Marshall A. G. (1998) A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass. Spectrom. 9, 225–233 [DOI] [PubMed] [Google Scholar]
- 12. Levin Y., Schwarz E., Wang L., Leweke F. M., Bahn S. (2007) Label-free LC-MS/MS quantitative proteomics for large-scale biomarker discovery in complex samples. J. Sep. Sci. 30, 2198–2203 [DOI] [PubMed] [Google Scholar]
- 13. Castagnola M., Inzitari R., Rossetti D. V., Olmi C., Cabras T., Piras V., Nicolussi P., Sanna M. T., Pellegrini M., Giardina B., Messana I. (2004) A cascade of 24 histatins (histatin 3 fragments) in human saliva: suggestions for a pre-secretory sequential cleavage pathway. J. Biol. Chem. 279, 41436–41443 [DOI] [PubMed] [Google Scholar]
- 14. Inzitari R., Cabras T., Onnis G., Olmi C., Mastinu A., Sanna M. T., Pellegrini M. G., Castagnola M., Messana I. (2005) Different isoforms and post-translational modifications of human salivary acidic proline-rich proteins. Proteomics 5, 805–815 [DOI] [PubMed] [Google Scholar]
- 15. Inzitari R., Cabras T., Rossetti D. V., Fanali C., Vitali A., Pellegrini M., Paludetti G., Manni A., Giardina B., Messana I., Castagnola M. (2006) Detection in human saliva of different statherin and P-B fragments and derivatives. Proteomics 6, 6370–6379 [DOI] [PubMed] [Google Scholar]
- 16. Lupi A., Messana I., Denotti G., Schininà M. E., Gambarini G., Fadda M. B., Vitali A., Cabras T., Piras V., Patamia M., Cordaro M., Giardina B., Castagnola M. (2003) Identification of the human salivary cystatin complex by the coupling of high-performance liquid chromatography and ion-trap mass spectrometry. Proteomics 3, 461–467 [DOI] [PubMed] [Google Scholar]
- 17. Messana I., Cabras T., Inzitari R., Lupi A., Zuppi C., Olmi C., Fadda M. B., Cordaro M., Giardina B., Castagnola M. (2004) Characterization of the human salivary basic proline-rich protein complex by a proteomic approach. J. Proteome Res. 3, 792–800 [DOI] [PubMed] [Google Scholar]
- 18. Messana I., Cabras T., Pisano E., Sanna M. T., Olianas A., Manconi B., Pellegrini M., Paludetti G., Scarano E., Fiorita A., Agostino S., Contucci A. M., Calò L., Picciotti P. M., Manni A., Bennick A., Vitali A., Fanali C., Inzitari R., Castagnola M. (2008) Trafficking and post-secretory events responsible for the formation of secreted human salivary peptides. A proteomic approach. Mol. Cell. Proteomics 7, 911–926 [DOI] [PubMed] [Google Scholar]
- 19. Marenholz I., Lovering R. C., Heizmann C. W. (2006) An update of S100 nomenclature. Biochim. Biophys. Acta. 1763, 1282–1283 [DOI] [PubMed] [Google Scholar]
- 20. Strupat K., Rogniaux H., Van Dorsselaer A., Roth J., Vogl T. (2000) Calcium-induced non covalently linked tetramers of MRP8 and MRP14 are confirmed by electrospray ionization-mass analysis. J. Am. Soc. Mass. Spectrom. 11, 780–788 [DOI] [PubMed] [Google Scholar]
- 21. Ohe Y., Hayashi H., Iwai K. (1989) Human spleen histone H1. Isolation and amino acid sequences of three minor variants, H1a, H1c, and H1d. J. Biochem. 106, 844–857 [DOI] [PubMed] [Google Scholar]
- 22. Heizmann C. W., Fritz G. (2009) The handbook of cell signaling, pp 943–994 Bradshaw RA, Dennis EA. Eds. CD ROM, Elsevier, Amsterdam [Google Scholar]
- 23. Leclerc E., Fritz G., Vetter S. W., Heizmann C. W. (2009) Binding of S100 proteins to RAGE: an update. Biochim. Biophys. Acta. 1793, 993–1007 [DOI] [PubMed] [Google Scholar]
- 24. Heizmann C. W., Ackermann G. E., Galichet A. (2007) Pathologies involving the S100 proteins and RAGE. Subcell. Biochem. 45, 93–138 [DOI] [PubMed] [Google Scholar]
- 25. Presland R. B., Dale B. A. (2000) Epithelial structural proteins of the skin and oral cavity: function in health and disease. Crit. Rev. Oral. Biol. Med. 11, 383–408 [DOI] [PubMed] [Google Scholar]
- 26. Fujimoto W., Nakanishi G., Arata J., Jetten A. M. (1997) Differential expression of human cornifin alpha and beta in squamous differentiating epithelial tissues and several skin lesions. J. Invest. Dermatol. 108, 200–204 [DOI] [PubMed] [Google Scholar]
- 27. Tesfaigzi J., Carlson D. M. (1999) Expression, regulation, and function of the SPR family of proteins. A review. Cell. Biochem. Biophys. 30, 243–265 [DOI] [PubMed] [Google Scholar]
- 28. Engelkamp D., Schäfer B. W., Mattei M. G., Erne P., Heizmann C. W. (1993) Six S100 genes are clustered on human chromosome 1q21: identification of two genes coding for the two previously unreported calcium binding proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 6547–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schäfer B. W., Wicki R., Engelkamp D., Mattei M. G., Heizmann C. W. (1995) Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics 25, 638–643 [DOI] [PubMed] [Google Scholar]
- 30. Robinson N. A., Lapic S., Welter J. F., Eckert R. L. (1997) S100A11, S100A10, annexin, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2 and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J. Biol. Chem. 272, 12035–12046 [DOI] [PubMed] [Google Scholar]
- 31. Robinson N. A., Eckert R. L. (1998) Identification of transglutaminase-reactive residues in S100A11. J. Biol. Chem. 273, 2721–2728 [DOI] [PubMed] [Google Scholar]
- 32. Mischke D., Korge B. P., Marenholz I., Volz A., Ziegler A. (1996) Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J. Invest. Dermatol. 106, 989–992 [DOI] [PubMed] [Google Scholar]
- 33. Turk V., Stoka V., Turk D. (2008) Cystatins: biochemical and structural properties, and medical relevance. Front. Biosci. 13, 5406–5420 [DOI] [PubMed] [Google Scholar]
- 34. Abrahamson M., Barrett A. J., Salvesen G., Grubb A. (1986) Isolation of six cysteine proteinase inhibitors from human urine. Their physicochemical and enzyme kinetic properties and concentrations in body fluids. J. Biol. Chem. 261, 11282–11289 [PubMed] [Google Scholar]
- 35. Zavasnik-Bergant T. (2008) Cystatin protease inhibitors and immune function. Front. Biosci. 13, 4625–4637 [DOI] [PubMed] [Google Scholar]
- 36. Rinne A., Dorn A., Järvinen M., Alavaikko M., Jokinen K., Hopsu-Havu V. K. (1986) Immunoelectron microscopical location of the acid cysteine proteinase inhibitor in the lymphatic tissue of the tonsils. Acta Histochem. 79, 137–145 [DOI] [PubMed] [Google Scholar]
- 37. van Nierop K., Muller F. J., Stap J., Van Noorden C. J., van Eijk M., de Groot C. (2006) Lysosomal destabilization contributes to apoptosis of germinal center B-lymphocytes. J. Histochem. Cytochem. 54, 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scott D. K., Lord R., Muller H. K., Malley R. C., Woods G. M. (2007) Proteomics identifies enhanced expression of stefin A in neonatal murine skin compared with adults: functional implications. Br. J. Dermatol. 156, 1156–1162 [DOI] [PubMed] [Google Scholar]
- 39. Ishida-Yamamoto A., Iizuka H. (1998) Structural organization of cornified cell envelopes and alterations in inherited skin disorder. Exp. Dermatol. 7, 1–10 [DOI] [PubMed] [Google Scholar]
- 40. Kos J., Lah T. (2006) Cystatins in cancer, pp. 153–165 Zerovnik E, Kopitar-Jerala N. (Ed.s) Human stefins and cystatins. Nova Biomedical Books, New York [Google Scholar]
- 41. Suzuki T., Hashimoto S., Toyoda N., Nagai S., Yamazaki N., Dong H. Y., Sakai J., Yamashita T., Nukiwa T., Matsushima K. (2000) Comprehensive gene expression profile in LPS-stimulated human monocytes by SAGE. Blood 96, 2584–2591 [PubMed] [Google Scholar]
- 42. Brännvall K., Hjelm H., Korhonen L., Lahtinen U., Lehesjoki A. E., Lindholm D. (2003) Cystatin-B is expressed by neural stem cells and by differentiated neurons and astrocytes. Biochem. Biophys. Res. Comm. 308, 369–374 [DOI] [PubMed] [Google Scholar]
- 43. Laitala-Leinonen T., Rinne R., Saukko P., Väänänen H. K., Rinne A. (2006) Cystatin B as an intracellular modulator of bone resorption. Matrix Biol. 25, 149–157 [DOI] [PubMed] [Google Scholar]
- 44. Sehnert B., Cavcic A., Böhm B., Kalden J. R., Nandakumar K. S., Holmdahl R., Burkhardt H. (2004) Antileukoproteinase: modulation of neutrophil function and therapeutic effects on anti-type II collagen antibody induced arthritis. Arthritis Rheum. 50, 2347–2359 [DOI] [PubMed] [Google Scholar]
- 45. Burkhardt H., Meyer P., Buchner E., Palombo-Kinne E., Hienzel-Wieland R., Becker W., Wolf F., Kalden J. R., Kinne R. W. (1997) The serine proteinase inhibitor antileukoproteinase specifically accumulates in normal but not in arthritic cartilage. J. Rheumatol. 24, 1145–1154 [PubMed] [Google Scholar]
- 46. Badamchian M., Damavandy A. A., Damavandy H., Wadhwa S. D., Katz B., Goldstein A. L. (2007) Identification and quantification of thymosin beta4 in human saliva and tears. Ann. N.Y. Acad. Sci. 1112, 458–465 [DOI] [PubMed] [Google Scholar]
- 47. Low T. L., Hu S. K., Goldstein A. L. (1981) Complete amino acid sequence of bovine thymosin β4: a thymic hormone that induces terminus deoxynucleotidyl transferase activity in thymocyte populations. Proc. Natl. Acad. Sci. U.S.A. 78, 1162–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hannappel E. (2007) β-Thymosins. Ann. N.Y. Acad. Sci. 1112, 21–37 [DOI] [PubMed] [Google Scholar]
- 49. Huff T., Müller C. S., Otto A. M., Netzker R., Hannappel E. (2001) β-Thymosins, small acidic peptides with multiple functions. Int. J. Biochem. Cell. Biol. 33, 205–220 [DOI] [PubMed] [Google Scholar]
- 50. Grant D. S., Kinsella J. L., Kibbey M. C., LaFlamme S., Burbelo P. D., Goldstein A. L., Kleinman H. K. (1995) Matrigel induces thymosin β4 gene in differentiating endothelial cells. J. Cell. Sci. 108, 3685–3694 [DOI] [PubMed] [Google Scholar]
- 51. Malinda K. M., Goldstein A. L., Kleinman H. K. (1997) Thymosin β4 stimulates directional migration of human umbilical vein endothelial cells. FASEB J. 11, 474–481 [DOI] [PubMed] [Google Scholar]
- 52. Smart N., Rossdeutsch A., Riley P. R. (2007) Thymosin β4 and angiogenesis: modes of action and therapeutic potential. Angiogenesis 10, 229–241 [DOI] [PubMed] [Google Scholar]
- 53. Smart N., Risebro C. A., Melville A. A., Moses K., Schwartz R. J., Chien K. R., Riley P. R. (2007) Thymosin β4 induces adult epicardial progenitor mobilization and neurovascularization. Nature 445, 177–182 [DOI] [PubMed] [Google Scholar]
- 54. Reti R., Kwon E., Qiu P., Wheater M., Sosne G. (2008) Thymosin beta 4 is cytoprotective in human gingival fibroblasts. Eur. J. Oral. Sci. 116, 424–430 [DOI] [PubMed] [Google Scholar]
- 55. Leclerc E., Heizmann C. W. (2010) The importance of the Ca2+/Zn2+ signaling S100 proteins and their receptor RAGE in translational medicine. Frontiers Biosci. in press [DOI] [PubMed] [Google Scholar]
- 56. Reya T., Morrison S. J., Clarke M. F., Weissman I. L. (2001) Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 [DOI] [PubMed] [Google Scholar]
- 57. Grichnik J. M. (2008) Melanoma, nevogenesis, and stem cell biology. J. Investig. Dermatol. 128, 2365–2380 [DOI] [PubMed] [Google Scholar]
- 58. Nemolato S., Restivo A., Cabras T., Zorcolo G., Di Felice E., Fanari M.U., Cau F., Gerosa C., Fanni D., Messana I., Castagnola M., Casula G., Faa G. (2010) Thymosin β4 in colorectal cancer is predominantly localized at the invasion front in tumor cells undergoing epithelial-mesenchymal transition. Eur. J. Cancer (submitted) [DOI] [PubMed] [Google Scholar]
- 59. Yang J., Weinberg R. A. (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14, 818–829 [DOI] [PubMed] [Google Scholar]