Abstract
Polo-like kinases regulate many aspects of mitotic and meiotic progression from yeast to man. In early mitosis, mammalian Polo-like kinase 1 (Plk1) controls centrosome maturation, spindle assembly, and microtubule attachment to kinetochores. However, despite the essential and diverse functions of Plk1, the full range of Plk1 substrates remains to be explored. To investigate the Plk1-dependent phosphoproteome of the human mitotic spindle, we combined stable isotope labeling by amino acids in cell culture with Plk1 inactivation or depletion followed by spindle isolation and mass spectrometry. Our study identified 358 unique Plk1-dependent phosphorylation sites on spindle proteins, including novel substrates, illustrating the complexity of the Plk1-dependent signaling network. Over 100 sites were validated by in vitro phosphorylation of peptide arrays, resulting in a broadening of the Plk1 consensus motif. Collectively, our data provide a rich source of information on Plk1-dependent phosphorylation, Plk1 docking to substrates, the influence of phosphorylation on protein localization, and the functional interaction between Plk1 and Aurora A on the early mitotic spindle.
During mitosis, multiple processes, such as mitotic entry, spindle assembly, chromosome segregation, and cytokinesis, must be carefully coordinated to ensure the error-free distribution of chromosomes into the newly forming daughter cells. The physical separation of the chromosomes to opposite poles of the cell is driven by the mitotic spindle, a proteinaceous and highly dynamic microtubule (MT)1-based macromolecular machine. Spindle assembly begins early in mitosis and is completed when the bipolar attachment of microtubules to kinetochore (KT) pairs is achieved (1, 2). Polo-like kinase 1 (Plk1), a serine/threonine-specific kinase first identified in Drosophila (3), is one of the key regulators of this essential mitotic process and has therefore attracted much attention (4–6). In agreement with its diverse functions, the localization of Plk1 during mitosis is dynamic. Plk1 first associates with centrosomes in prophase before it localizes to spindle poles and KTs in prometaphase and metaphase. During anaphase, Plk1 is recruited to the central spindle and finally accumulates at the midbody during telophase. Proteomics studies using oriented peptide libraries have shown that two so-called polo boxes at the C-terminal end of Plk1, the polo box domain (PBD), are crucial for the localization of this kinase to cellular structures (7, 8). This domain binds to specific phosphorylated sequence motifs that are created by other priming kinases or are self-primed by Plk1 itself, thus providing an efficient mechanism to regulate localization and substrate selectivity in time and space (9–11).
Despite the pleiotropic and critical functions of Plk1 during mitosis, only a limited number of target proteins and phosphorylation sites on substrates have so far been identified or studied in detail (4–6, 12). The difficulties in identification of bona fide Plk1 substrates stem from the low abundance of some substrates, technical limitations for determining in vivo phosphorylation sites, the requirement for Plk1 localization for recognition of some substrates, and the possibility that Plk1 may phosphorylate a broader consensus motif than determined previously (13). Recent developments in mass spectrometry (MS)-based proteomics have allowed the identification of a large number of in vivo phosphorylation sites from complex samples (14). However, the nature of the kinase(s) responsible for most of these phosphorylation events is still unclear, and the assignment of phosphorylation sites to individual kinases remains a challenging task. Previously, we explored the human mitotic spindle by MS and successfully identified a large number of novel spindle proteins and phosphorylation sites (15, 16). Now, the development of quantitative methods to monitor in vivo phosphorylation changes in complex samples (17–19) represents a unique opportunity to address the role of individual kinases in spindle function.
To study Plk1 function at the mitotic spindle, we combined quantitative proteomics using stable isotope labeling by amino acids in cell culture (SILAC) (20) with the isolation of human mitotic spindles and phosphopeptide enrichment. To expand the experimental coverage of Plk1 substrates and gain further insight into direct and indirect functions of Plk1, we compared the phosphoproteomes of mitotic spindles isolated from cells lacking Plk1 activity with spindles from cells with fully active kinase. Two independent approaches were used to interfere with Plk1 activity: protein depletion using an inducible small hairpin (shRNA) cell line and selective inhibition of the kinase by the small molecule inhibitor ZK-thiazolidinone (TAL) (21). Phosphorylation sites found to be down-regulated after Plk1 inhibition/depletion were subsequently validated using in vitro phosphorylation of synthetic peptide arrays. This approach identified many candidate Plk1 substrates, allowed confirmation of direct phosphorylation by Plk1 of more than 100 sites identified in vivo, and suggested a broader phosphorylation consensus motif for this kinase. Collectively, our data set provides a rich resource for in-depth studies on the spindle-associated Plk1-dependent phosphoproteome. This is illustrated by selective follow-up studies in which we validated the Plk1-dependent localization of substrates to centrosomes and kinetochores. In particular, using a phosphospecific antibody, we confirmed Plk1-dependent CENP-F phosphorylation in vivo and demonstrated that CENP-F localization to kinetochores depends on Plk1 kinase activity. Furthermore, we identified several Aurora A-dependent phosphorylation events that are regulated by Plk1, supporting the emerging view of an intimate functional relationship between Plk1 and Aurora A kinase (22, 23).
EXPERIMENTAL PROCEDURES
Cell Culture, SILAC Media, Synchronization, and Spindle Isolation
For SILAC, HeLa S3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) deficient in amino acids arginine and lysine and supplemented with 5% dialyzed FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, and either unlabeled arginine·HCl and lysine·HCl (SILAC light) or l-[U-13C6,15N4]arginine·HCl and l-[U-13C6,15N2]lysine· HCl (SILAC heavy) (Cambridge Isotope Laboratories) at concentrations of 42 (arginine) and 72 μg/ml (lysine). Cells were grown at 37 °C in a humidified incubator with 5% atmospheric CO2. Cells were adapted to the appropriate SILAC medium for at least six passages to achieve complete incorporation of the isotope-labeled amino acids. For large scale mitotic spindle isolation, each population of HeLa S3 labeled cells was propagated to five triple flasks with a total surface of 500 cm2. Cells were first presynchronized with thymidine (2 mm) for 20 h, then washed twice with PBS, and released from the thymidine block into TAL or monastrol (MA)-containing medium (1 or 150 μm, respectively) or by induction with 1 μg/ml tetracycline for 36 h in the case of the inducible cell lines. Taxol-stabilized mitotic spindles (including kinetochores and centrosomes) were isolated as described previously (24).
In-gel Protein Digestion
Enriched spindle proteins were separated by SDS-PAGE and in-gel (25) digested with trypsin. In each experiment, about 400 μg of the spindle fraction was loaded to one NuPAGE Bis-Tris gel and run for 50 min using a 200-V/100-mA program. Gels were stained with a 1:1 mixture of 0.2% Coomassie Blue in methanol (MeOH) and 20% acetic acid for 40 min and destained using a mixture of 30% MeOH and 10% acetic acid for 15 min, 10% acetic acid for 1 h, and 2% acetic acid overnight at 4 °C. Gels were cut into 20 slices each. After reduction and alkylation, proteins were digested by 15 ng/μl trypsin for 16 h. Digested peptides were extracted with 30% ACN and 5% formic acid and dried.
Phosphopeptide Enrichment and Desalting
Phosphorylated peptides were selectively enriched by titanium dioxide beads with lactic acid as a modifier (26). A piece of C8 material was plugged at the constricted end of a GELoader tip, and about 3 mg of titanium dioxide beads was transferred to each of the microcolumns. The microcolumns were washed with 40 μl of 0.3 μg/μl lactic acid in a mixture of 80% ACN and 0.2% TFA. Digested peptides were redissolved in 0.3 μg/μl lactic acid in a mixture of 80% ACN and 2% TFA and applied to microcolumns with a slow flow rate, allowing phosphorylated peptides to bind to the titanium dioxide beads. The microcolumns were subsequently washed with 40 μl of 0.3 μg/μl lactic acid in a mixture of 80% ACN and 0.2% TFA and 40 μl of a mixture of 80% ACN and 0.2% TFA. Phosphorylated peptides were eluted with 40 μl of 0.6% NH4OH and 40 μl of a mixture of 80% ACN and 0.2% TFA. The eluates were dried and redissolved in 0.5% formic acid for LC-MS/MS analysis. The flow-through fractions were desalted with C18 reversed-phase material prior to LC-MS/MS for protein expression level measurement. Briefly, a piece of C18 material was plugged into a GELoader tip and washed with 20 μl of 2-propanol and 20 μl of 5% formic acid. The dried flow-through fractions were redissolved in 20 μl of 5% formic acid and applied to the C18 microcolumns. The columns were then washed with 20 μl of 5% formic acid, and subsequently peptides were eluted with 2 × 20 μl of 50% methanol and 2% formic acid. The eluates were dried and redissolved in 0.5% formic acid for LC-MS/MS.
Nano-LC-MS/MS Analysis
The nano-LC-MS/MS analysis was performed with a nanoACQUITY ultraperformance liquid chromatography (UPLC) system (Waters) connected to a hybrid linear ion trap/orbitrap tandem mass spectrometer (Thermo Electron). Dissolved peptides were loaded at 500 nl/min into a pulled and fused silica capillary with an inner diameter of 75 μm and a tip of 8 μm (New Objective) packed to a length of 12 cm with reversed-phase ReproSil-Pur C18-AQ 3-μm resin (Maisch) and eluted at 200 nl/min by a stepwise 180-min gradient of 0–100% buffer A (0.2% formic acid in water) and buffer B (0.2% formic acid in acetonitrile). The linear ion trap/orbitrap (LTQ-Orbitrap) mass spectrometer was operated in a data-dependent MS/MS mode. Survey full-scan MS spectra (from m/z 300 to 2000) were acquired in the orbitrap with a resolution of 60,000 at m/z 400. A maximum of five peptides were sequentially isolated for fragmentation in the linear ion trap using collision-induced dissociation (CID). The lock mass option was enabled to improve mass accuracy as described (27). All spectra were acquired with Xcalibur software.
Data Processing and Analysis
MS spectra were searched via MASCOT (28) (version 2.2.0, Matrix Science, London, UK). Searches were performed against the International Protein Index (IPI) human (version 3.48; 71,400 protein entries) database that was concatenated with decoy database sequences (142,800 protein entries in total) (29). Peak lists were generated using MaxQuant (30) (version 1.0.12.5) with the following parameters: top six MS/MS peaks for 100 Da, three data points for centroid, Gaussian centroid determination, slice peaks at local minima. MaxQuant identifies potential SILAC pairs based on characteristics such as mass differences of labeled amino acids and intensity correlation over elution time to establish peptide peak lists. MaxQuant was also used for SILAC quantitation, false discovery rate (FDR) determination, peptide/protein grouping, phosphorylation site localization probability scoring, and data filtering based on MASCOT search results. The initial precursor mass tolerance was set to ±7 ppm, whereas an accuracy of ±0.5 Da was used for MS/MS fragmentation spectra. Enzyme specificity was set to trypsin, allowing cleavages N-terminal to proline. Carbamidomethylation was set as fixed modification. Oxidation, protein N-terminal acetylation, Arg10, Lys8, and phosphorylation (Ser/Thr/Tyr) were considered as variable modifications. A maximum of three labeled amino acids and two missed tryptic cleavages and a minimum peptide length of six amino acids were allowed. A minimum of one unique peptide was required for protein identification. In the case where the set of identified peptides in one protein was equal to or completely contained in the set of identified peptides of another protein, these two proteins were joined in the same protein group. Shared peptides are most parsimoniously associated within the group of the protein with the highest number of identified peptides but remain in all groups where they occur (30). The protein with the highest number of identified peptides is presented as the leading protein in the supplemental tables with the highest confidence in its protein group. To ensure the fidelity of protein and phosphorylation site identifications, the target-decoy database strategy was used to minimize FDRs in our data set (29). A composite database is created by a “target” protein sequence database appropriate to the protein mixture to be analyzed (IPI human version 3.48), and a “decoy” database is created by reversing the target protein sequences. Because the reverse transformation preserves amino acid frequencies, protein and (approximate) peptide length distributions, and approximate mass distributions of theoretical peptides, the number of spectra matching decoy sequences gives an estimate of FDR. Estimated peptide/protein FDR was calculated as follows: (Number of hits in the reversed database/Number of hits in the forward database) × 100% (31). A maximum 5% peptide FDR and 2% protein FDR were allowed for the first step peptide/protein identification. A MASCOT ion score of at least 12 and phosphorylation site localization probability equal to or higher than 75% were used for data filtering. However, we included in a separate table (supplemental Table S5, A and B) all down-regulated phosphorylation sites (see below) with lower MASCOT scores (7 ≤ x < 12) identified from inhibitor (supplemental Table S5A) and shRNA (supplemental Table S5B) experiments as some of these sites might also be functionally relevant and may be considered for validation by in vitro spotting experiments (see below) despite their lower confidence. Automated quantitation was accomplished by the MaxQuant software. To correct for possible changes of protein levels on the mitotic spindle, the SILAC ratio of each phosphorylated peptide was normalized to the SILAC ratio of the corresponding protein (calculated as the mean of all unmodified peptide ratios from this protein).
To compare the protein levels on isolated spindles with those in total lysates by MS (see Fig. 5A), ratios (H/L: TAL/MA) on the spindle were normalized against the average ratio of all α/β-Tubulin subunits identified to correct for variability in spindle isolation (supplemental Table S4A). Next, the ratios from the two spindle experiments (biological replicates) were averaged, and proteins with ratios that followed a different trend between biological replicates were excluded (above and below 1 ± 0.2). To define the relative change in spindle localization, the averaged normalized ratio for each protein from the spindle preparation experiment was divided by the corresponding normalized ratio from total lysates.
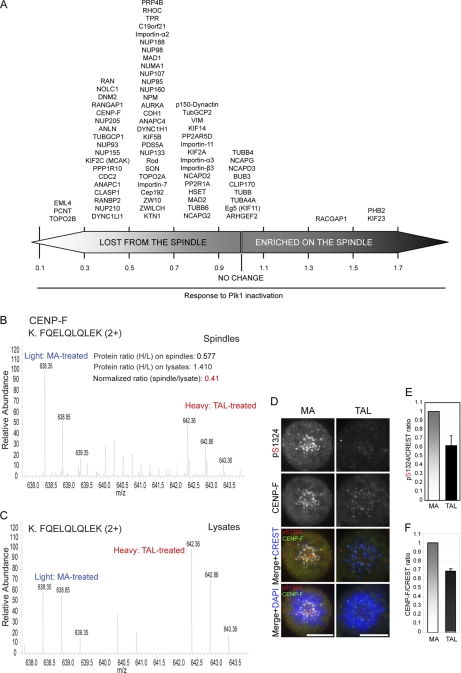
Fig. 5.
Plk1-dependent localization of spindle components. A, proteins are arranged according to the effect of Plk1 inactivation on their association with the spindle. Ratio of 1 indicates no change on spindle levels relative to amounts in total lysate (see supplemental information and supplemental Table S4). B and C, representative MS spectra of a CENP-F phosphopeptide quantified from spindles (B) and total lysates (C) of MA/TAL-treated cells. D, HeLa S3 cells treated with MA or TAL were fixed and immunostained for Ser(P)-1324 CENP-F (red), CENP-F (green), and CREST (far red; depicted in blue). DNA was visualized using DAPI (blue). Scale bars, 10 μm. E and F, fluorescence intensity of Ser(P)-1324 CENP-F and total CENP-F, respectively, was quantified on KTs in MA- or TAL-treated cells. Intensities were plotted after normalization for CREST staining (10 cells/>10 KTs per cell, n = 3 independent experiments). Columns and error bars show average and S.D., setting a ratio of 1 for MA-treated samples.
The orbitrap raw data (MS raw files), MS/MS peak lists, and MS/MS spectra associated with this study were submitted to the Tranche data repository and can be downloaded from https://proteomecommons.org/tranche/ using the following hash and pass phrase: Santamariaetal: zt4vKJ7TYsdufcxOekHmhXJAmlNxac61TD8lMT/CWcE6LaOVcB/no0hnzd2ws3cZrvmApH65svbnb0QtkFuc0T85GtkAAAAAACJdhQ==.
Bioinformatics
Motif-X was used with a significance cutoff of 0.0001 and at least 10 occurrences of the motif in the set. IPI human was used as a background data set with the built-in function. STRING 8.0 was queried with human protein symbols from supplemental Table S2, A and B. Confidence was set to medium (0.400), and only options “Databases” and “Experiments” were selected. Conservation of phosphorylated residues was checked with BLAST searches against rat (RGSC3.4.54), mouse (NCBIM37.54), zebrafish (Zv8.54), Drosophila melanogaster (BDGP5.4), Xenopus tropicalis (JGI4.1.54), Caenorhabditis elegans (WS190.54), and Saccharomyces cerevisiae (SGD1.0) proteomes obtained from the Ensembl FTP server. First, close sequence homologs of the proteins were identified using an E-value cutoff of 1e−50. Then, the phosphorylated site was checked for exact conservation in the alignment.
Peptide Spot Array Synthesis and Phosphorylation Assay
Peptide arrays were constructed using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on a MultiPep robotic spotter according to the manufacturer's directions (Intavis). For Plk1 assays on peptide-immobilized cellulose membranes, dried membranes were first washed in ethanol and then hydrated in kinase buffer (50 mm Tris-HCl, pH7.5, 10 mm MgCl2, 1 mm DTT, 100 μm NaF, and 10 μm sodium orthovanadate) for 1 h followed by overnight blocking in kinase buffer with 100 mm NaCl and 0.5 mg/ml BSA. The next day, the membrane was blocked again with kinase buffer containing 1 mg/ml BSA, 100 mm NaCl, and 10 μm cold ATP at 30 °C for 45 min. The block was subsequently replaced with kinase reaction buffer containing 0.2 mg/ml BSA, 50 μCi/ml [γ-32P]ATP (3000 Ci/mmol, 10 mCi/ml), 150 nm recombinant His-Plk1, and 10 μm ATP for 3 h on a shaker at 30 °C. The membranes were then washed extensively (10 × 15 min in 1 m NaCl, 3 × 5 min in H2O, 3 × 15 min 5% H3PO4, and 3 × 5 min in H2O) and then sonicated overnight in 8 m urea, 1% SDS (w/v), and 0.5% (v/v) β-mercaptoethanol to remove residual nonspecific radioactivity. The membranes were washed again with H2O followed by ethanol and dried before being visualized by autoradiography. Aurora A assays were performed similarly but using Aurora A kinase buffer (20 mm Hepes, pH 7.4, 150 mm KCl, 5 mm MnCl2, 5 mm NaF, and 1 mm DTT). Sites were considered positive when differential signals could be observed between the Ser/Thr/Tyr and the Ala version of the peptide, negative when no detectable signal was observed, and non-conclusive when the peptide was also phosphorylated on the membrane incubated with ATP alone as control or alternatively when the signal between the Ser/Thr/Tyr and the Ala version of the peptide was indistinguishable.
RESULTS
Experimental Strategy to Compare Spindle Phosphoproteomes in Presence or Absence of Plk1 Activity
To investigate the Plk1-dependent phosphoproteome of human mitotic spindles, we used a quantitative phosphoproteomics strategy that combines SILAC with selective enrichment of spindle-associated proteins. To enable quantitation of the changes of phosphopeptide abundance by MS, cells were labeled by growing them in medium containing either normal arginine and lysine (Arg0/Lys0) or the heavy isotopic variants [13C6,15N4]arginine and [13C6,15N2]lysine (Arg10/Lys8) (20) (Fig. 1A). To synchronize control cells at a similar mitotic stage and, importantly, to obtain similar microtubule arrays as in Plk1-inactivated cells, we interfered with the function of the kinesin motor protein Eg5. Like Plk1 inactivation (32), Eg5 inactivation results in a prometaphase arrest with a monoastral microtubule array surrounded by a ring of chromosomes (33). Importantly, Plk1 localization and kinase activity were retained in Eg5-inactivated cells (Figs. 2, A and G, and 6, C and D; see also supplemental Fig. S1C and “Discussion”) (21), comparable with prometaphase cells arrested with nocodazole or Taxol (Fig. 2H).
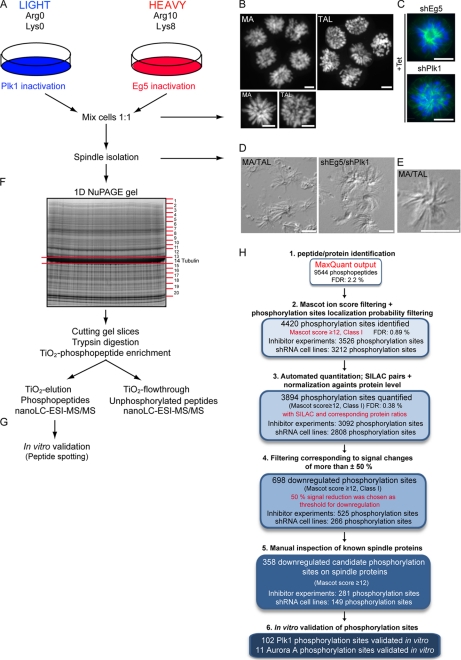
Fig. 1.
Strategies to study Plk1 phosphoproteome on mitotic spindles. A, schematic of the design of SILAC experiments. HeLa S3 cells grown in the presence of light or heavy isotope-labeled arginine and lysine were subjected to Tet induction or MA/TAL treatment, respectively. In three experiments, cells for Plk1 or Eg5 inactivation were grown in light or heavy SILAC medium, respectively, whereas the fourth experiment (Inhibitor-1) was performed under a reverse labeling condition. B, DNA (4′-6-diamidino-2-phenylindole, DAPI staining) is shown for MA- and TAL-treated cells. C, DNA and spindle morphology is shown for shEg5- and shPlk1-induced cells (α-Tubulin in green; DNA in blue). D, DIC pictures of isolated mitotic spindles from MA/TAL-treated cells (left panel) and shEg5/shPlk1-depleted cells (right panel). E, DIC picture from a single monopolar spindle from a mixture of MA/TAL-treated cells. Scale bars, 10 μm. F, analytical strategy to map phosphorylation sites in spindle proteins. G, validation of direct Plk1 phosphorylation sites by in vitro Plk1 kinase assays on peptide arrays. H, flowchart indicating the initial MaxQuant phosphopeptide output (raw data) and the subsequent filtering of candidates, ending with the validation in vitro: MaxQuant output (supplemental Table S1A), Class I phosphorylation sites identified with MASCOT score ≥12, quantified sites (with SILAC pair and normalized against protein ratios), down-regulated sites, down-regulated sites on spindle proteins tested in vitro, and Plk1 and Aurora A sites on spindle proteins validated in vitro.
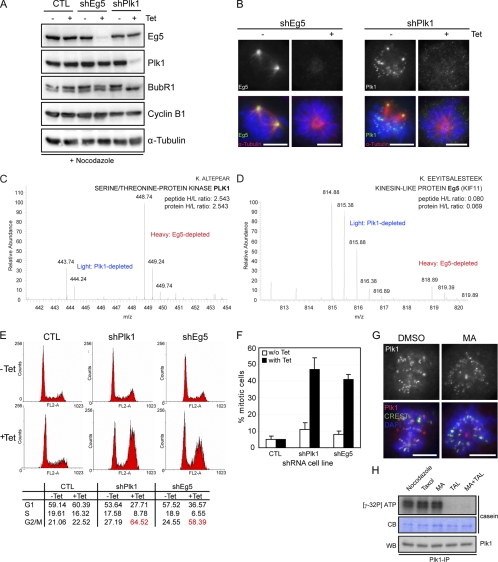
Fig. 2.
Establishment of stable inducible shRNA cell lines for Plk1 or Eg5 depletion. A, lysates from control (CTL), shEg5, and shPlk1 cells treated for 36 h with Tet or left for 36 h in Tet-free medium and treated for the last 12 h with nocodazole were used for Western blotting analysis. Membranes were probed for Eg5, Plk1, BubR1, Cyclin B1, and α-Tubulin as loading control. B, shEg5 and shPlk1 cells treated for 36 h with Tet or left for 36 h in Tet-free medium were fixed and stained with the indicated antibodies. Eg5 and Plk1 are shown in green, and α-Tubulin is shown in red. DNA was visualized using DAPI (blue). C and D, mass spectra representing a SILAC peak pair for Plk1 and Eg5, respectively, from shEg5/shPlk1-induced cells. Note that for Plk1 (C) we could only identify one SILAC unphosphorylated peptide pair. E, FACS analysis was performed from control, shPlk1, and shEg5 cells treated as in B. The percentage of cells with 2N (G1 and S) or 4N (G2/M) content is shown. F, mitotic index from control, shEg5, and shPlk1 cells treated for 36 h with Tet or left in Tet-free medium (300 cells for each condition; n = 3). Error bars represent standard deviation. G, DMSO- and MA-treated cells were fixed and stained for the indicated antibodies. Plk1 is shown in red, and CREST is shown in green. DNA was visualized using DAPI (blue). Scale bars, 10 μm. H, Plk1 in vitro kinase assay. Plk1 was immunoprecipitated from nocodazole-, Taxol-, MA-, TAL-, and MA + TAL-treated cells, and casein was used as substrate. Protein phosphorylation was visualized by autoradiography of 32P, equal amounts of casein are shown by Coomassie Blue (CB), and immunoprecipitates were probed by Western blotting (WB) with anti-Plk1 antibody.
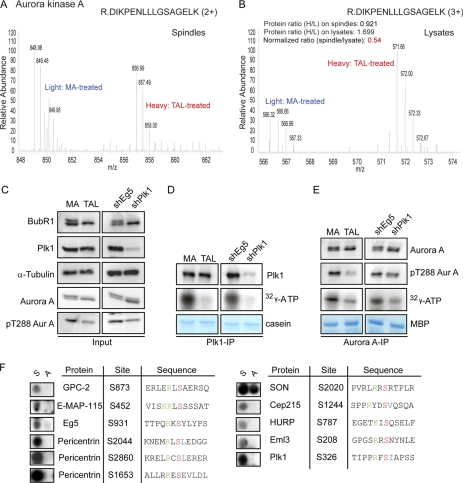
Fig. 6.
Reduced Aurora A activity as consequence of Plk1 inactivation. A and B, representative MS spectra of an Aurora A peptide quantified from spindles (A) and total lysates (B) of MA/TAL-treated cells. C–E, Western blotting for the indicated proteins in cell lysates from MA/TAL-treated (left panels) or shEg5/shPlk1 cells (right panels) (C). In vitro kinase assays using Plk1 (D) or Aurora A (Aur A) (E) are shown. Kinases were immunoprecipitated from MA/TAL-treated or shEg5/shPlk1-induced cells, and casein (D) and myelin basic protein (MBP) (E) were used as substrates. Protein phosphorylation was visualized by autoradiography of 32P. Immunoprecipitates (D and E) were probed by Western blotting for the indicated proteins (F). Phosphorylation sites on Aurora A substrates were validated by in vitro kinase assays on peptide arrays as described in the legend to Fig. 4C.
To disrupt Plk1 activity (or Eg5 activity for control), two experimental strategies were used. First, we interfered with protein function by lowering protein levels. To this end, we generated tetracycline (Tet)-inducible stable HeLa S3 cell lines (shPlk1 and shEg5 for control), allowing the shRNA-mediated knockdown of Plk1 and Eg5, respectively. After shRNA induction, Eg5 and Plk1 protein levels were efficiently reduced when compared with non-induced or control cells (a stable cell line generated with an empty vector) (Fig. 2, A and B). As expected, depletion of Plk1 but not Eg5 led to loss of BubR1 hyperphosphorylation (Fig. 2A), previously shown to be Plk1-dependent (34–36). Analysis by MS of the protein levels of Plk1 and Eg5 after Tet induction of the shRNA cell lines provided an independent assessment for depletion efficiency (Fig. 2, C and D). Flow cytometry (FACS) analysis of Tet-induced shEg5 or shPlk1 cells revealed an accumulation of cells with 4N DNA content, indicative of a G2/M arrest (Fig. 2E). Concomitantly, a striking increase in the mitotic index was observed (Fig. 2F) in agreement with the predominant phenotype seen upon interference with Plk1 or Eg5 (32, 33).
As a second, complementary approach, we disrupted Plk1 kinase activity using the small molecule inhibitor TAL (21). MA, a small molecule inhibitor of Eg5 (37), was used to treat control cells. Both inhibitors are highly specific toward their targets. TAL showed high selectivity for Plk1 when tested against a panel of 93 kinases with only two close relatives, Plk2 and Plk3, displaying significant sensitivity to TAL, and it produced cellular phenotypes that were indistinguishable from those caused by other, structurally unrelated Plk1 inhibitors (5, 21). MA, on the other hand, was shown to specifically inhibit Eg5-driven microtubule motility, whereas it had no effect on processes controlled by other motor proteins (37).
Taxol-stabilized mitotic spindles (containing KTs and centrosomes) were purified essentially as described previously (24). To ensure minimal variation in sample handling and processing, equalized amounts of Plk1- and Eg5-depleted/inhibited cells were combined prior to spindle isolation (Fig. 1, A–C). Examination of the purified spindles by differential interference contrast (DIC) light microscopy revealed predominantly monopolar spindles (Fig. 1, D and E), as expected, largely free of other cellular structures. Purified spindles were subsequently solubilized and separated by one-dimensional gradient SDS-PAGE. Gel slices covering the entire protein mass range were digested with trypsin, and the resulting peptide fractions were subjected to phosphopeptide enrichment on TiO2 beads. Both TiO2 elution and TiO2 flow-through fractions (phosphorylated and unphosphorylated peptides, respectively) were then analyzed using a nano-LC-ESI orbitrap (Fig. 1F). Two independent experiments were performed for each Plk1 inhibition strategy, providing biological replicates (see below). To address the reproducibility of phosphorylation site identification and quantitation, samples from one of the experiments performed with the Tet-inducible stable cell lines were split, separated on distinct one-dimensional gels, and analyzed as technical replicates. A good reproducibility (r2 value of 0.84) was observed (supplemental Fig. S1A) in line with other phosphoproteome studies (18, 38).
Plk1-dependent (Phospho)proteome
Data from four independent experiments (two experiments performed with small molecule inhibitors and two performed with shRNA cell lines) resulted in a total of 3894 unique phosphorylation sites (MASCOT score cutoff of ≥12 (supplemental Table S1, B and C); Class I sites according to Ref. 17) for which SILAC and protein ratios could be established. This number included 3092 and 2808 sites from experiments performed with small molecule inhibitors and the shRNA cell lines, respectively (52% overlap in identified sites; see supplemental Fig. S2A). Fig. 1H shows a flowchart of all filtering and experimental validation steps performed in this study. Detailed information on the numerical analysis of phosphopeptides and phosphorylation sites is provided as supplemental information.
To choose optimal thresholds for defining up- and down-regulated phosphorylation sites, we analyzed a 1:1 mixture of identically treated but differentially labeled mitotic lysates (where TAL was applied to both SILAC light and heavy labeled HeLa S3 cells; see supplemental Fig. S1B). We found that only 1.9% of the detected peptides had SILAC ratios >3/2, whereas 4.4% were detected with ratios <2/3. According to the initial SILAC labeling used in the experiments performed with small molecule inhibitors or shRNA cell lines (i.e. Inhibitor-2 and shRNA experiments; H/L: MA/TAL and Inhibitor-1; H/L: TAL/MA), the error rate considering the selected thresholds for the down-regulated sites was below 2% in three of our experiments and around 4% only in one experiment. Because our work flow included a subsequent in vitro kinase assay for the verification of potential Plk1 targets, we considered these error rates as a good compromise between experimental sensitivity and stringency and defined 3/2 and 2/3 (corresponding to signal changes of more than ±50%) as thresholds for up- and down-regulation, respectively. Consistent with this definition, a change of 50% was also considered significant for regulated phosphorylation events in other recent SILAC-based phosphoproteomics studies (39–42). Phosphorylation sites were considered as down-regulated according to the following criteria. All sites showing contradictory behavior (down-regulated in one experiment but up-regulated in the biological replicate) were discarded, whereas sites that were down-regulated in one experiment but unchanged (ratios between 2/3 and 3/2) or undetectable in the replicate were retained in the analysis. Data obtained using the two different strategies, shRNA-mediated depletion or small molecule-induced inhibition, were analyzed similarly (supplemental Table S2, A and B). However, in this case, even contradictory data sets were retained for further biological analysis. In total, 698 Class I phosphorylation sites in 304 different proteins were down-regulated in at least one of the four experiments, 525 sites were down-regulated after TAL treatment, and 266 sites were down-regulated after Plk1 depletion by shRNA (supplemental Table S2, A and B, and Fig. 1H; for one example of MS spectra, see supplemental Fig. S3). In addition, we also evaluated our data using a “two-standard deviation” cutoff. Filtering the list of 3894 identified phosphorylation sites using this criterion resulted in a very similar set of down-regulated phosphorylation sites (709 sites compared with 698 using the 50% change criterion) with a very high overlap (90.4%), further validating the chosen cutoff. In our data set, the maximum observed down-regulation (in the protein Nesprin-1) corresponds to ratios of 0.0005 and 0.0027 for inhibitor and shRNA experiments, respectively. On average, a 2.25-fold change in phosphorylation was observed in the down-regulated phosphorylation sites.
Plk1 Spindle Substrates
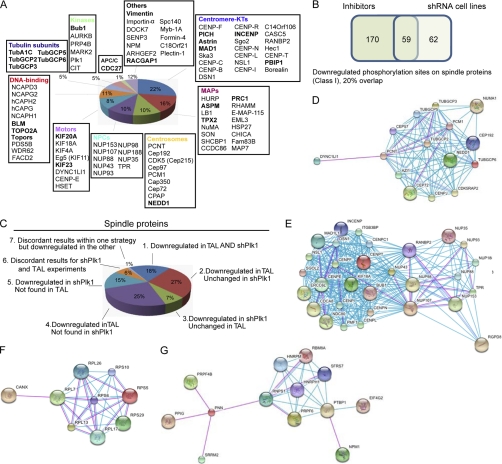
We estimate that 101 of the 304 human proteins (33%) with Plk1-dependent phosphorylation sites identified here have known mitotic spindle functions (supplemental Table S3, A and B) (15, 16), demonstrating a substantial enrichment for spindle components in our samples. In total, 358 phosphorylation sites were down-regulated on spindle proteins upon Plk1 inactivation either by TAL treatment or stable shPlk1-mediated depletion (supplemental Table S3, A and B). In agreement with essential functions of Plk1 at the spindle poles, phosphorylation sites of centrosomal, MT-associated proteins (MAPs), and motor proteins were identified to be down-regulated upon Plk1 inactivation (Fig. 3A). Among the spindle proteins containing down-regulated sites, those associated with the centromere or KT structure were overrepresented, consistent with an important role for Plk1 in KT function (Fig. 3A). This included the centromeric protein PBIP1, the protein kinase Bub1, and the subunit of the chromosomal passenger complex, INCENP, all shown to contribute to KT localization of Plk1 during mitosis (9, 43, 44). Interestingly, members of the nuclear pore complexes (NPCs), shown previously to associate with KTs at the onset of mitosis (45), also exhibited down-regulated sites after Plk1 inhibition (Fig. 3A). Finally, other mitotic kinases, tubulin subunits, and proteins functioning in chromosome structure during mitosis also appeared to be regulated by Plk1. Our data set contained several known Plk1 substrates, such as Cdc27, INCENP, Kif20A, and topoisomerase IIA (5, 46), validating our approach (Fig. 3A).
Fig. 3.
Plk1-dependent substrates. A, pie chart depicting proteins with down-regulated phosphorylation sites upon Plk1 inactivation. Proteins are classified according to function and localization. Known Plk1 substrates are shown in bold. B, Venn diagram illustrating the overlap between down-regulated phosphorylation sites (Class I) identified by shRNA and inhibitor experiments on spindle proteins. C, pie chart depicting the regulation of phosphorylation sites on spindle proteins upon Plk1 inhibition/depletion. Sites were classified according to whether they were down-regulated, unchanged, or not found in shPlk1- and/or TAL-treated cells. D–G, subnetworks generated using the STRING database (78) with all proteins containing down-regulated phosphorylation sites upon Plk1 inactivation (see supplemental Fig. S5 for complete interaction network). D and E, centrosomal and KT-NUP subnetworks identified only upon TAL treatment. F and G, ribosome-binding protein subnetworks. A medium confidence setting (0.400) was used, and only experimental information (pink) or information from databases (light blue) was used. APC/C, anaphase-promoting complex/cyclosome.
We also evaluated the set of spindle proteins with down-regulated sites (supplemental Table S3, A and B) for the presence of S(pS/pT)P (where pS is phosphoserine and pT is phosphothreonine) motifs as it has been previously shown that phosphopeptides bind to the Plk1-PBD through such motifs (7). Sixty-six of the 101 (65%) spindle proteins with Plk1-dependent phosphorylation sites contained such a predicted PBD recognition motif. However, we note that a similar percentage (62%) of all proteins from the complete human IPI, when matched for comparable average length, also contained at least one such potential PBD recognition motif (supplemental Fig. S4A). This suggests that additional features may be required to rigorously predict a functional PBD in a full-length Plk1 substrate. Importantly, our study identified the vast majority (∼80%) of spindle proteins carrying both a PBD-docking site and putative Plk1 phosphorylation site(s) identified in a previous independent study on Plk1-PBD binding partners (10). However, only half of these proteins showed down-regulation at Plk1 phosphorylation sites (supplemental Fig. S4B). Interestingly, the other half comprised proteins involved in late mitotic processes, notably Anilin, Rock2, Septin, and Cdh1, which would suggest an absence of Plk1-dependent regulation on these proteins during early mitosis. We also compared our data set with a study on the identification of substrates of Cdc5, the budding yeast homolog of Plk1 (47). However, of the five bioinformatically predicted substrates validated by chemical genetics, none of the putative human orthologs exhibited Plk1-regulated phosphorylation in our study (data not shown).
We also compared the results on the regulation of phosphorylation sites as obtained from the two different Plk1 inactivation strategies. Our data show that chemical inhibition of Plk1 resulted in a larger number of down-regulated phosphorylation sites and a stronger down-regulation at individual sites than Plk1 depletion (Fig. 3, B and C, supplemental Fig. S2B, and supplemental Table S3A). Most likely this reflects both incomplete depletion of Plk1 in the shRNA cell line and the very potent inhibition of Plk1 achieved by use of TAL (21). Surprisingly, although the total number of down-regulated sites identified upon Plk1 depletion was lower, around half of these were not identified in the TAL-treated sample (Fig. 3B). This strongly argues in favor of using different strategies to interfere with Plk1.
To detect protein-protein networks regulated by Plk1, we asked whether putative Plk1 substrates were enriched for members of particular interaction networks using the STRING database (see supplemental Fig. S5A for complete interaction network and “Experimental Procedures”). In agreement with an enrichment of spindle components, two large subnetworks, including centrosomal as well as a kinetochore and nuclear pore complex proteins, appeared enriched among putative Plk1 substrates regardless of the strategy used for their identification (Fig. 3, D and E). Interaction network analyses were also performed separately for the putative substrates identified through TAL or shPlk1 experiments. Although the analysis of the shPlk1 data set did not reveal additional subnetworks of apparent functional relevance, in the group of phosphoproteins down-regulated only upon Plk1 inhibition, we observed two interaction networks containing ribosome-associated and mRNA-binding proteins (Fig. 3, F and G). To complete this analysis, we also asked whether substrates of other members of the Plk family were present among the phosphoproteins identified in this study. We could not identify any STRING-predicted interaction partners for Plk2 or Plk3 in our data set (not shown) but found several putative Plk4 interaction partners (supplemental Fig. S5B, left panel). However, these proteins were also predicted by STRING to be associated with Plk1 (supplemental Fig. S5B, right panel). These latter results support the interpretation that TAL- or shPlk1-induced down-regulation of phosphorylation on the proteins identified here reflects that action of Plk1 rather than any other Plk family member.
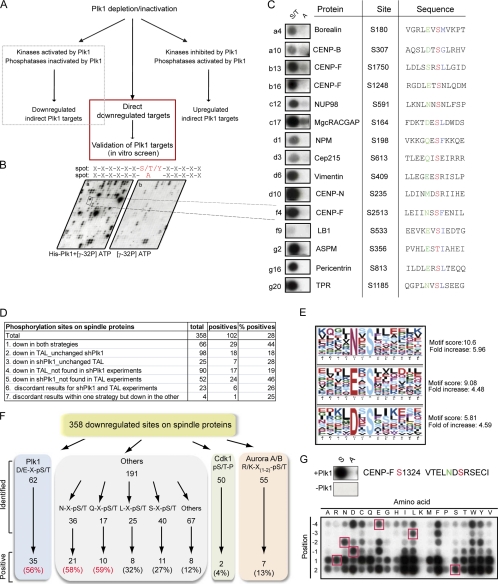
Validation of Plk1-dependent Phosphorylation Events on Spindle Proteins through a Candidate-based in non capital Vitro Screen
As a consequence of reduced Plk1 activity, phosphorylation sites on direct Plk1 substrates are expected to be down-regulated in Plk1-depleted or TAL-treated cells. However, considering that kinases and phosphatases often form complex signaling networks, the pool of regulated phosphorylation sites is also expected to include indirect targets of Plk1 activity, notably proteins that are targets of kinases or phosphatases that act downstream of Plk1 (Fig. 4A). Only 17% of the down-regulated sites on spindle proteins conformed to the previously described Plk1 consensus motif (D/E)X(S/T) (13), whereas 14 and 15% matched the consensus motifs for Cdk1 and Aurora A/B, respectively (Fig. 4F). To distinguish direct from indirect substrates of Plk1 activity, we adopted a peptide phosphorylation approach to validate potential direct Plk1 target sites. Using peptides synthesized directly on cellulose membranes as substrates, we performed in vitro Plk1 kinase assays with recombinant Plk1 purified from insect cells (Figs. 1G and 4B). The 358 down-regulated sites identified on spindle components were arrayed on the membrane as 12-mer peptides with the potential phosphoacceptor amino acid kept constant at position 7 in either its serine/threonine/tyrosine native form or changed to alanine to verify signal specificity (Fig. 4B). Incubation of duplicate peptide arrays (generated simultaneously) with either kinase and [32P]ATP or [32P]ATP alone followed by extensive washing resulted in clear and specific signals on membranes phosphorylated by Plk1, whereas only a few spots were detected on control membranes incubated with [32P]ATP alone (Fig. 4, B and C, and supplemental Fig. S6A) (see “Experimental Procedures”). In total, 102 down-regulated sites could be phosphorylated in vitro by Plk1 using this approach (Table I and supplemental Table S3, A and B; see also Fig. 4C for examples). More than 60% of these sites appeared to be conserved in at least three (human and two other) species selected for comparison, indicating that many of the detected phosphorylation sites may be functionally relevant (Table I and supplemental Table S3, A and B). Indeed, functionally relevant residues recently shown to be phosphorylated by Plk1, such as Ser-170 on MgcRacGAP (48), scored positive using this approach (Table I).
Fig. 4.
Validation of Plk1 phosphorylation sites. A, schematic representing direct and indirect consequences of Plk1 inhibition. B, example of the in vitro approach to validate Plk1 phosphorylation sites. C, examples of phosphorylation sites on validated Plk1 substrates. Shown are (from left to right) peptide spots (phosphoacceptor (S/T) to the left; alanine substitution (A) to the right), protein name, phosphorylated residue, and target peptide. Phosphoacceptor residues are marked in red, −2 positions are marked in green, and hydrophobic residues at +1 position are marked in blue. D, table summarizing in vitro results on the number of down-regulated phosphorylation sites on spindle proteins. Only the differences between the percentage of positive sites responding exclusively to TAL (18 or 19%) and down-regulated in both strategies (44%) were statistically significant (Fisher's test, p value <0.05). E, Motif-X analysis of validated Plk1 phosphorylation sites on spindle proteins. F, schematic showing the number of identified and validated down-regulated sites on spindle proteins, displayed according to different motifs. G, permutational analysis for Plk1 phosphorylation on the CENP-F peptide containing Ser-1324; residues at positions −4 to +2 were substituted by every other amino acid. Red squares mark the positions of the original amino acid in the peptide; S and A refer to peptides with the original amino acid acceptor or the alanine substitution.
Table I. Validated Plk1 phosphorylation sites on known spindle proteins.
Known Plk1 substrates and Plk1 phosphorylation site are marked in italic, and conserved sites are marked in bold (species are specified in supplemental Table S3A, column P).
| Proteins | Accession number | Phosphorylation sites |
|---|---|---|
| ARHGEF2 | Q92974 | Ser-737 |
| ASPM | Q8IZT6 | Thr-178, Ser-267, Ser-270, Ser-280, Ser-355, Thr-356, Ser-1825, Ser-3426 |
| AURKB | Q96GD4 | Tyr-239 |
| BLM | P54132 | Ser-26,a Ser-28a |
| Borealin | Q53HL2 | Ser-180 |
| BUB1 | O43683 | Ser-661 |
| CASC5 | Q8NG31 | Ser-1013, Ser-1808 |
| CDC27 | P30260 | Ser-441 |
| CENP-B | P07199 | Ser-306, Ser-307 |
| CENP-C | Q03188 | Ser-96, Ser-104, Ser-110, Thr-516, Ser-250, Thr-734 |
| CENP-E | Q02224 | Ser-611, Ser-612 |
| CENP-F | P49454 | Ser-242, Ser-838, Ser-1248, Ser-1324, Ser-1750, Ser-1988, Ser-2512, Ser-2513 |
| CENP-I | Q92674 | Ser-709 |
| CENP-J (CPAP) | Q9HC77 | Ser-556a |
| CENP-L | Q8N0S6 | Ser-53 |
| CENP-N | Q96H22 | Ser-226, Ser-235, Ser-282 |
| CENP-T | Q96BT3 | Ser-45 |
| CENPU (PBIP1) | Q71F23 | Ser-194 |
| C14Orf106 | Q6P0N0 | Ser-135, Ser-192 |
| Cep97 | Q8IW35 | Ser-308 |
| Cep170 | Q5SW79 | Ser-880a |
| Cep192 | Q8TEP8 | Ser-1502 |
| CDK5 (Cep215) | Q96SN8 | Ser-613, Ser-1102 |
| Cep350 | Q5VT06 | Ser-2689 |
| DGL7 (HURP) | Q86T11 | Ser-777 |
| ERCC6L (PICH) | Q2NKX8 | Ser-774, Ser-790 |
| INCENP | Q9NQS7 | Ser-72, Ser-330 |
| KIF18A | Q8NI77 | Ser-681a |
| KIF20A | O95235 | Tyr-558, Ser-635 |
| KIF23 | Q02241 | Ser-867,aThr-897, Ser-889 |
| KIFC1 (HSET) | Q9BW19 | Ser-33 |
| KIF4A | O95239 | Ser-394, Ser-815, |
| LB1 | Q8WWK9 | Ser-533 |
| MAD1 | Q9Y6D9 | Ser-8,a Ser-490a |
| NCAPD3 | P42695 | Thr-430, Ser-508 |
| NCAPG2 | Q86XI2 | Ser-30, Thr-1114 |
| NCAPH | Q15003 | Thr-98 |
| NPM | P06748 | Ser-198 |
| NuMA | Q14980 | Thr-1818 |
| NUP88 | Q99567 | Ser-540 |
| NUP93 | Q8N1F7 | Ser-72 |
| NUP98 | P52948 | Ser-591 |
| NUP107 | P57740 | Ser-4, Ser-57 |
| NUP153 | P49790 | Ser-343 |
| PCM1 | Q15154 | Ser-110 |
| PCNT | O95613 | Ser-813, Ser-815, Thr-1688, Thr-1690, Ser-2594, Thr-3325, Ser-3326 |
| PRC1 | O43663 | Ser-554, Ser-592 |
| RACGAP1 | Q9H0H5 | Ser-164, Ser-170, Ser-214 |
| SGOL2 | O562F6 | Ser-436, Ser-1151a |
| SON | P18583 | Ser-154 |
| SPAG5 (ASTRIN) | Q96R06 | Ser-401 |
| Sperm-specific antigen 2 | P28290 | Ser-737, Ser-739 |
| TOP2A | P11388 | Ser-282 |
| TPR | P12270 | Ser-1185 |
| TPX2 | Q9ULW0 | Ser-356, Thr-361, Ser-654 |
| TUBA1C | Q9BQE3 | Ser-41, Ser-48 |
| TUBGCP5 | Q96RT8 | Ser-182 |
| Vimentin | P08670 | Ser-83, Ser-409, Thr-458, Ser-459 |
| WD62 | Q9UFV9 | Ser-974 |
| ZWINT | O95229 | Ser-84a |
a Sites that were identified with high confidence in earlier experiments (analyzed with MSQuant) but only reached a final MASCOT score <12 in this study (and hence were not included in supplemental Table S3A).
Of the phosphorylation sites that were down-regulated in vivo regardless of the strategy used for Plk1 inactivation, 44% scored positive in the in vitro kinase assay. A similar proportion of sites responding to Plk1 shRNA could be validated (28 or 46%, depending on whether sites were unchanged or not found upon TAL treatment, respectively; Fig. 4D). In contrast, of the sites responding exclusively to TAL, only 18 or 19% (unchanged or not found upon shPlk1, respectively) scored positive in the in vitro assay (Fig. 4D). Taken at face value, this indicates that strong and rapid inactivation of Plk1 by TAL is more likely to reveal indirect effects than the relatively slow and incomplete inactivation achieved by shRNA.
Reconsideration of Plk1 Consensus Motif
Analysis of the in vitro validated Plk1 sites using Motif-X (79) revealed that the previously proposed Plk1 kinase consensus motif (D/E)X(S/T)Φ (where X is any amino acid and Φ is a hydrophobic amino acid) (13) applies to only a subset of validated substrates (Fig. 4, C and F, and supplemental Table S3A). Although 56% of peptides conforming to this consensus scored positive in the in vitro kinase assay, similarly high success rates were observed for peptides that carry an asparagine (Asn) or glutamine (Gln) at position −2 (Fig. 4F, supplemental Table S3A, and supplemental Experimental Procedures for statistics). Furthermore, Asn at position −2 was also overrepresented in the Motif-X analysis (Fig. 4E), consistent with the frequent identification of corresponding peptides in our data set as well as previous reports of Asn at the −2 position in substrates of Plk1 (or its yeast homolog Cdc5p) in the literature, including the anaphase-promoting complex/cyclosome subunit Cdc27 (49), the cohesion subunit Rec8 (50), the cytokinesis effector kinase Rock2 (10), and the centralspindlin complex component MgcRacGAP (48). To further explore the optimal Plk1 consensus motif, we used peptide arrays to examine the tolerance of Plk1 for different amino acids at several positions relative to the phosphoacceptor residue. Specifically, we chose the sequence surrounding CENP-F Ser-1324 as it contained an Asn in the −2 position, conformed well to the Motif-X consensus, and was clearly down-regulated in both the Plk1 shRNA and Plk1 inhibitor studies (Fig. 4G and supplemental Table S3A). Using this peptide as a starting point, each residue from positions −4 to +2 was substituted with every other natural amino acid, and the ability of Plk1 to phosphorylate the resulting peptides was analyzed in an in vitro phosphorylation assay (Fig. 4G). Incubating a duplicate control membrane with [32P]ATP in the absence of Plk1 resulted in virtually no signal (supplemental Fig. S6B). In the context of this peptide, our results demonstrate a strong preference of Plk1 for particular residues N-terminal to the phosphoacceptor amino acid. In contrast, most amino acid substitutions were well tolerated at the +1 and +2 position, although there was also a clear preference for hydrophobic residues. One notable exception was proline at the +1 position (Fig. 4G), which was not tolerated. This strongly suggests that (S/T)P motifs are not substrates for Plk1. Furthermore, we again find marked preferences for Asn or Glu at the −2 position, whereas Asp was not strongly preferred over other residues, and, within the context of this particular peptide, Gln was not well tolerated (Fig. 4G). This clearly shows that Glu/Asp/Asn/Gln are not interchangeable in all Plk1 substrate peptides. According to a similar analysis performed on six different peptides carrying an Asn at the −2 position, we conclude that some peptides accept Glu/Asp but not Gln, whereas others accept Gln but not Glu/Asp (supplemental Fig. S7). Significantly, a leucine in position −3 was prominent in many validated phosphopeptides (Fig. 4, C and G, and supplemental Table S3A), and substitution of this residue was poorly accepted in the permutation analysis performed on the CENP-F peptide (Fig. 4G). Collectively, these observations are in agreement with both our MS data and the results of the in vitro peptide phosphorylation analyses, and they prompt us to propose that the consensus sequence for Plk1 can be broadened to L(Φ)(E/N/D(Q))X(S/T)L(Φ). We emphasize, however, that even this broadened consensus does not account for all observed Plk1 substrates.
Plk1-dependent Localization of Spindle Components
In accordance with the requirement for Plk1 activity to properly localize some of its substrates on the mitotic spindle (5), we considered it possible that the localization of some downstream targets would be altered as a consequence of Plk1 inhibition. To analyze this possibility in a systematic and quantitative manner, we treated cells with either TAL or MA, mixed the samples, and then determined protein ratios for spindle preparations as well as total lysates from identically treated cells (supplemental Table S4A; see also supplemental Table S4, B and C, for MaxQuant protein identification output from spindle and lysates experiments, respectively, and “Experimental Procedures”). Proteins showing reduced or increased association with the spindle apparatus are displayed in Fig. 5A (note that only proteins with SILAC ratios identified in both spindle and total lysate experiments are shown). Interestingly, many proteins that showed down-regulated phosphorylation sites upon Plk1 inactivation were also depleted from the spindle (Fig. 3A and supplemental Table S3, A and B), arguing strongly that Plk1 activity is required for their spindle association.
An intriguing case was CENP-F, a large coiled coil protein that transiently localizes to outer KTs and preferentially to KTs of unaligned chromosomes (51). Although it was previously shown that CENP-F exhibits slower electrophoretic mobility upon mitotic entry (52), little is known about CENP-F phosphorylation. We identified 19 serine and threonine phosphorylation sites on CENP-F that were suppressed by more than 2-fold upon Plk1 inhibition, eight of which we could validate in vitro (Table I and supplemental Table S3, A and B). Our MS data further suggest that the levels of CENP-F on the spindle apparatus were reduced upon Plk1 inactivation when compared with the corresponding levels in total lysates (Fig. 5, A–C, and supplemental Table S4A). In agreement with this observation, immunofluorescence analysis revealed a significant reduction of total CENP-F and CENP-F Ser-1324 phosphorylation signal at KTs upon TAL treatment (Fig. 5, D–F). Phosphorylation at Ser-1324 was monitored using a phosphospecific antibody generated toward this residue (see supplemental Fig. S8 for antibody characterization and specificity). These data demonstrate that CENP-F is a likely Plk1 target and that CENP-F localization is regulated by Plk1. In addition, they confirm in vivo phosphorylation by Plk1 of sites conforming to the broadened consensus motif defined above.
Consequences of Plk1 Inactivation on Centrosomal Substrates
Plk1 inhibition reduces the amounts of Plk1 associated with centrosomes (21, 35). Although centrosomes display functionality even after disruption of the centrosomal localization of this kinase (53, 54), the recruitment of Plk1-dependent centrosomal components is expected to be impaired. Strikingly, levels of pericentrin and Cep192 were clearly attenuated in the spindle preparations upon TAL treatment (Fig. 5A). In addition, we observed decreased Cep215 levels on the spindles of Plk1-inactivated cells (although we failed to detect Cep215 in total cell lysates for normalization) (supplemental Table S4A). We also identified and validated several phosphorylation sites on pericentrin, Cep192, and Cep215 (Table I) and confirmed by immunofluorescence analysis that all three proteins revealed defective centrosome accumulation upon Plk1 inactivation (supplemental Fig. S9, A–C; see also Ref. 55). In contrast, the localization of another centrosomal protein, Cep135, that did not display down-regulated phosphorylation sites and hence served as a control was indistinguishable between DMSO- (control), MA-, TAL-, or MA + TAL-treated cells (supplemental Fig. S9D). These data strongly argue that pericentrin, Cep192, and Cep215 are novel Plk1 substrates, and they support an independent study identifying these proteins as Plk1-regulated components involved in the recruitment of γ-Tubulin complexes to mitotic centrosomes (55).
Integration of Aurora A and Plk1 Activity
Aurora A localization to centrosomes has recently been shown to depend on Plk1 activity (21, 53, 56). In agreement with this observation, our MS data show that Aurora A levels on the isolated spindle were reduced by 46% upon Plk1 inhibition when normalized against total lysates (Figs. 5A and 6, A and B). It has also recently been postulated that Plk1 contributes to efficient Aurora A activation (57, 58). Indeed, compared with immunoprecipitates from control (Eg5-inhibited) cells, both Plk1 and Aurora A showed reduced in vitro kinase activity when immunoprecipitated from Plk1-inhibited cells (Fig. 6, C–E). Although the reduction in activity was expected for Plk1, the significant reduction in activity observed in parallel for Aurora A is consistent with the notion that inhibition of Plk1 also reduces Aurora A activity. In further support of the above conclusion, we emphasize that several of the 358 phosphorylation sites identified as down-regulated after Plk1 depletion/inhibition by MS analysis conform to the Aurora A/B consensus motif (Fig. 4F) (59, 60). Interestingly, many of these sites were present in proteins likely to be Aurora A substrates, including known Aurora A interactors, such as TPX2 (61) or centrosomal proteins and MAPs involved in centrosome maturation and spindle assembly (supplemental Table S3C). To corroborate a direct role of Aurora A in the phosphorylation of these substrates, Aurora A kinase assays were performed in vitro on peptide arrays in the same manner as described above for Plk1. Included in this assay were all phosphopeptides that (i) were down-regulated upon Plk1 inactivation but negative in the Plk1 in vitro assays, (ii) conformed to an Aurora consensus motif, and (iii) were potential Aurora A substrates according to their reported function and localization (see supplemental Table S3C). These experiments revealed six novel candidate Aurora A substrates and 11 new Aurora A phosphorylation sites (Fig. 6F and Table II). Collectively, the data support the idea that Aurora A is an important downstream effector of Plk1 activity.
Table II. Validated Aurora A phosphorylation sites on known spindle proteins.
Known Aurora A substrates are marked in italics.
DISCUSSION
Phosphoproteomics has emerged as a powerful approach for the study of complex regulatory networks (19, 62–64). In addition to providing comprehensive inventories of phosphorylation sites, proteomics approaches based on either isotope labeling techniques or label-free quantitation methods offer the opportunity for studying quantitative aspects of intracellular signaling. In this study, we deployed quantitative proteomics methods in conjunction with the inhibition of Plk1, a major regulatory kinase known to be essential for cell cycle progression through mitosis. Our data provide insight into the phosphoproteome that is controlled, directly or indirectly, by Plk1. Furthermore, we used in vitro peptide spotting assays and recombinant Plk1 for in vitro phosphorylation analysis to validate many of the potential Plk1 target sites inferred from our quantitative proteomics data set. These latter results led us to propose that physiological Plk1 target sites conform to a broader consensus sequence than hitherto recognized.
Complementary Strategies to Explore the Plk1-dependent Phosphoproteome
Despite the increased understanding of Plk1 function in recent years (4–6, 65), many of its physiological functions cannot be explained by the substrates identified to date. Here, we used a combination of approaches to extend our knowledge of Plk1 signaling. Through the isolation of the spindle apparatus prior to analysis by mass spectrometry, we were able to focus our study specifically on Plk1 targets and phosphorylation events related to mitotic centrosomes, spindle poles, and spindle-associated proteins. This approach, in combination with an efficient TiO2-based phosphopeptide enrichment protocol and high resolution orbitrap mass spectrometry, allowed for the detection and SILAC-based quantitation of almost 4000 phosphorylation sites. Because protein phosphorylation states often regulate spindle localization, it was important to normalize all phosphorylation ratios to the corresponding changes in protein levels. In this context, the prior reduction in sample complexity by spindle isolation was crucial for recovering sufficient numbers of unphosphorylated peptides (from non-TiO2 enriched samples) for normalization to protein levels. Of particular importance, we used two different but complementary experimental strategies for identification of Plk1 targets, namely inhibition of kinase activity by the small molecule inhibitor TAL and depletion of endogenous Plk1 through use of a stable inducible shRNA cell line. Collectively, we identified many previously uncharacterized downstream targets and new Plk1-specific in vivo phosphorylation sites, many of which could be validated in vitro.
Although 52% of the total 3894 phosphorylation sites could be identified through both experimental strategies, the phosphorylation sites identified in biological replicates of TAL inhibition or shPlk1 depletion experiments revealed 38 and 32% overlap, respectively (supplemental Fig. S2A). These variabilities may be explained in part by differences in spindle purity and variability during the phosphoenrichment process and MS procedure. In addition, they are likely to reflect differences in the efficiency and/or kinetics of Plk1 inactivation in response to the two strategies (inhibition versus depletion). It is striking that many more Plk1-dependent sites were found to be regulated in response to TAL treatment than shPlk1-mediated depletion. Likewise, the extent of down-regulation was often more pronounced in the former case. Only 13% of all sites could be shown to be down-regulated in both approaches (supplemental Fig. S2B). This low degree of overlap may appear surprising, but we emphasize that both experimental and biological reasons concur to reduce this number (for a review, see Ref. 66). On the experimental side, shRNA is not expected to be sufficiently effective to reveal down-regulation of all phosphopeptides over the time course of the experiment; conversely, TAL treatment is likely to have been so efficient that in some cases no residual phosphopeptide was available for determination of a SILAC ratio. From the biological perspective, several reasons might explain regulation observed upon TAL treatment but not after shRNA-mediated depletion. First, we note that more putative Cdk1 and Aurora A substrates were found to be affected by TAL treatment than by shPlk1, whereas in vitro validation of putative Plk1 targets revealed a smaller percentage of positive hits in the case of TAL-responsive sites. This suggests that rapid and thorough inhibition of Plk1 by TAL facilitates the detection of indirect downstream targets. Second, it is possible that some TAL-sensitive phosphorylation sites reflect the action of Plk2, Plk3, or unknown protein kinases, although the latter is unlikely (21). Third, some high affinity Plk1 sites may show only minor down-regulation in response to TAL, particularly if phosphatases fail to remove the corresponding phosphates within the time frame of our experiments. Finally and most interestingly, we note that some phosphopeptides were down-regulated only in response to shPlk1 but not TAL treatment. Taken at face value, phosphorylation of these peptides would thus seem to depend on the presence of Plk1 protein rather than Plk1 activity. However, this conclusion suffers from the caveat that this latter class of phosphopeptides also includes sites for which no SILAC pair could be identified in the TAL experiments.
A priori, it could be argued that some of the observed changes in phosphorylation levels might reflect increased phosphorylation in response to Eg5 inactivation (in the samples analyzed for control) rather than decreased phosphorylation upon Plk1 inactivation. It is formally possible that Eg5 depletion or inhibition affects the activity of kinases and/or the availability of substrates, but at present, no such effects are described in the literature. We measured the in vitro activities of several mitotic kinases, notably Plk1, Aurora A, Aurora B, Cdk1, and Nek2, and found none of these to be affected by Eg5 depletion or inhibition (shown for Plk1 in Fig. 2H; not shown for the other kinases). Likewise, we showed that Plk1 localization is not affected by Eg5 depletion or inhibition (Fig. 2G). Furthermore, label-free quantitation revealed strong correlation of the regulation of sites responding to TAL treatment alone with that seen upon combined MA + TAL treatment, indicating that the bulk of regulation is due to Plk1 inhibition (and not up-regulation in response to Eg5 inhibition; supplemental Fig. S1C). Most importantly, many of the putative Plk1-regulated phosphorylation events were subsequently validated by in vitro kinase assays. Thus, although we cannot rigorously exclude that a few of the described substrates are actually hyperphosphorylated in response to Eg5 inactivation (rather than dephosphorylated upon Plk1 inactivation), we are confident that this explanation cannot possibly account for the bulk of the results.
Validation of Putative Plk1 Spindle Targets and Broadening of Plk1 Consensus Motif
Our analysis revealed 358 phosphorylation sites on spindle proteins that were down-regulated upon Plk1 inhibition/depletion, but surprisingly, only 17% of these sites matched the classical Plk1 consensus motif ((E/D)X(pS/T)) (13). A study combining peptide arrays with in vitro kinase assays led us to propose a broadened consensus that also favors Leu at position −3 and allows Asn or Gln at position −2. Although this broadened consensus accommodates many more peptides scoring positive for in vitro phosphorylation by Plk1, we emphasize that in vivo specificity of Plk1 is certainly influenced by parameters that cannot be represented in a linear recognition motif. Another interesting feature emerging from a global analysis of the proteins containing validated Plk1 phosphorylation sites is that 30% of these did not contain a PBD recognition motif conforming to the consensus S(pS/T)P. This suggests that a third of all Plk1 substrates involve Plk1 docking through sites either that are primed by kinases other than Cdk1 or that are not primed at all.
Plk1 Regulation of Centrosome-associated Proteins
In agreement with a role of Plk1 in spindle assembly and function, many proteins appeared to be mislocalized from the spindle upon Plk1 inactivation. This may reflect a direct regulation by phosphorylation or may be an indirect effect of the role of Plk1 in controlling spindle dynamics (67). For example, we observed a reduction of spindle-associated KIF2A upon Plk1 inactivation in agreement with a recent report (68). Furthermore, we found that subunits of the γ-Tubulin ring complex as well as proteins implicated in γ-Tubulin recruitment to the centrosome (the centrosomal proteins pericentrin, Cep192, and Cep215 (55)) were also lost from spindles upon Plk1 inhibition. One striking example for cooperation between mitotic kinases may be seen at the G2 to M transition when Plk1 cooperates with Aurora A to induce centrosome maturation (4, 6, 69). In particular, it has recently been proposed that Plk1 regulates Aurora A activity and vice versa (57, 58, 70, 71). Here we identified a subset of phosphorylation sites that showed down-regulation upon Plk1 inhibition but conformed to Aurora A sites and, moreover, could readily be phosphorylated by Aurora A in vitro. These results lend strong support to the proposal that the role Plk1 in centrosome maturation may be explained, at least in part, by its impact on Aurora A localization and activity (57). We also identified substrates common to both Plk1 and Aurora A, such as pericentrin, the γ-Tubulin subunit GCP2, and the centrosomal protein Cep215, confirming that these two kinases cooperate to control key early mitotic functions through co-regulation of several important substrates.
Plk1 Regulation of Kinetochore Proteins
We identified novel phosphorylation sites down-regulated after Plk1 inhibition on INCENP, Bub1, Hec1, and PIBP1 (also known as CENP-U (9)). Plk1-dependent phosphorylation of these substrates would be consistent with a role of these proteins in targeting Plk1 to KTs; in addition, however, they may play an important role in regulating KT functions independently of Plk1 recruitment. Once Plk1 is bound to KTs, little is known about key Plk1 substrates at this site. We identified down-regulated sites upon Plk1 inactivation on proteins known to contribute to the establishment and/or maintenance of KT-MT interactions (such as the KNL-1/Mis12 complex/Ndc80 complex (KMN) network and Ska components; see Fig. 3A) (1). In addition, we validated phosphorylation sites sensitive to Plk1 inhibition on CENP-F and CENP-E, both of which are required for proper chromosome congression. Our MS analysis also revealed reduced Mad1/Mad2 levels on the spindle as well as down-regulated phosphorylation sites on Mad1 upon Plk1 inactivation. Although Plk1 is not essential for the spindle assembly checkpoint in somatic cells (5), decreased levels of Mad1 and Mad2 at the kinetochores have been previously reported upon Plk1 depletion (9, 72). Our results are in agreement with these findings and suggest a possible role of Plk1 in fine tuning the localization of several checkpoint components. Collectively, our study reveals several novel Plk1-dependent phosphorylation events that are likely to contribute to control centrosome maturation, spindle assembly, and kinetochore function.
Novel Plk1-regulated Components of Spindle?
Interestingly, a number of NPC components, including several nucleoporins (NUPs), were also identified as putative Plk1 substrates. It is possible that members of the NUP family are regulated by Plk1, perhaps controlling complex formation or KT localization after nuclear envelope breakdown (45). NPC proteins, notably the Nup107–160 complex, have recently been implicated in microtubule nucleation from KTs (73) and in the regulation of complex formation between the translocated promoter region and the spindle assembly checkpoint components Mad1 and Mad2 (74). A subset of ribosome-associated and RNA-binding proteins was also enriched among the putative Plk1 substrates. Whether the presence of these proteins in our spindle preparations (see also Ref. 16) reflects contamination or a genuine spindle association remains to be clarified. In support of a physiological role of RNA-binding proteins in the spindle, we note that conserved classes of mRNA were found to be enriched on this structure (75), and PRPF4, a protein kinase implicated in the regulation of mRNA splicing (also identified in our data set), has been shown to localize to KTs during mitosis (76). It is tempting, therefore, to speculate that Plk1 activity contributes to the association of these proteins with the spindle apparatus perhaps to ensure their proper segregation during cell division. Another possibility is that Plk1 contributes to suppress cap-dependent translation and/or enhance cap-independent translation during mitosis (77). In support of this latter notion, we observed down-regulation of phosphorylation sites on two members of the eIF4F complex upon Plk1 inhibition.
In summary, our comprehensive study of phosphoregulation by Plk1 vastly extends the inventories of direct Plk1 substrates, Plk1-dependent phosphorylation sites on the mitotic spindle apparatus, and downstream effectors of Plk1 in mammalian cells. These data should thus serve as a valuable resource for future research in the field. The strategies developed here for the analysis of the Plk1-dependent proteome and phosphoproteome on the mitotic spindle can in principle be applied to other protein kinases provided that effective shRNA duplexes and/or specific inhibitors are available. These strategies thus respond to a growing contemporary need for assigning experimental phosphoproteome data sets to specific individual kinases.
Acknowledgments
We thank Jürgen Cox and Matthias Mann for early access to the MaxQuant program. We gratefully acknowledge Albert Ries for technical assistance and Kalyan Dulla, Thorsten Schmidt, and other members of the Nigg laboratory for insightful discussions and helpful suggestions.
Footnotes
* This work was supported by the Max Planck Society as well as by Experimental Network for Functional Integration Contract LSHG-CT-2005-518254 funded by the European Commission within its FP6 Program.
 This article contains supplemental Figs. S1–S9, Tables S1–S5, and Experimental Procedures.
This article contains supplemental Figs. S1–S9, Tables S1–S5, and Experimental Procedures.
1 The abbreviations used are:
- MT
- microtubule
- Plk
- Polo-like kinase
- KT
- kinetochore
- PBD
- polo box domain
- SILAC
- stable isotope labeling by amino acids in cell culture
- TAL
- ZK-thiazolidinone
- MA
- monastrol
- LTQ
- linear triple quadrupole
- IPI
- International Protein Index
- FDR
- false discovery rate
- DIC
- differential interference contrast
- MAP
- MT-associated protein
- NPC
- nuclear pore complex
- NUP
- nucleoporin
- CENP
- centromere protein
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- H/L
- heavy/light
- Tet
- tetracycline
- INCENP
- inner centromere protein
- MgcRacGAP
- male germ cell Rac GTPase-activating protein
- CREST
- calcinosis, raynaud phenomenon, esophageal dismotility, sclerodactyly, telangectlasia
- DAPI 4′
- 6-diamidino-2-phenylindole
- shRNA
- small hairpin RNA.
REFERENCES
- 1. Cheeseman I. M., Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 2. Musacchio A., Salmon E. D. (2007) The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 3. Llamazares S., Moreira A., Tavares A., Girdham C., Spruce B. A., Gonzalez C., Karess R. E., Glover D. M., Sunkel C. E. (1991) polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 5, 2153–2165 [DOI] [PubMed] [Google Scholar]
- 4. Barr F. A., Silljé H. H., Nigg E. A. (2004) Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 5. Petronczki M., Lénárt P., Peters J. M. (2008) Polo on the rise—from mitotic entry to cytokinesis with Plk1. Dev. Cell 14, 646–659 [DOI] [PubMed] [Google Scholar]
- 6. Archambault V., Glover D. M. (2009) Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10, 265–275 [DOI] [PubMed] [Google Scholar]
- 7. Elia A. E., Cantley L. C., Yaffe M. B. (2003) Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 8. Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. (2003) The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95 [DOI] [PubMed] [Google Scholar]
- 9. Kang Y. H., Park J. E., Yu L. R., Soung N. K., Yun S. M., Bang J. K., Seong Y. S., Yu H., Garfield S., Veenstra T. D., Lee K. S. (2006) Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell 24, 409–422 [DOI] [PubMed] [Google Scholar]
- 10. Lowery D. M., Clauser K. R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S. E., Gammeltoft S., Carr S. A., Yaffe M. B. (2007) Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 26, 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neef R., Preisinger C., Sutcliffe J., Kopajtich R., Nigg E. A., Mayer T. U., Barr F. A. (2003) Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Plyte S., Musacchio A. (2007) PLK1 inhibitors: setting the mitotic death trap. Curr. Biol. 17, R280–283 [DOI] [PubMed] [Google Scholar]
- 13. Nakajima H., Toyoshima-Morimoto F., Taniguchi E., Nishida E. (2003) Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 278, 25277–25280 [DOI] [PubMed] [Google Scholar]
- 14. Schreiber T. B., Mäusbacher N., Breitkopf S. B., Grundner-Culemann K., Daub H. (2008) Quantitative phosphoproteomics—an emerging key technology in signal-transduction research. Proteomics 8, 4416–4432 [DOI] [PubMed] [Google Scholar]
- 15. Nousiainen M., Silljé H. H., Sauer G., Nigg E. A., Körner R. (2006) Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U.S.A. 103, 5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sauer G., Körner R., Hanisch A., Ries A., Nigg E. A., Silljé H. H. (2005) Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics 4, 35–43 [DOI] [PubMed] [Google Scholar]
- 17. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 18. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 20. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 21. Santamaria A., Neef R., Eberspächer U., Eis K., Husemann M., Mumberg D., Prechtl S., Schulze V., Siemeister G., Wortmann L., Barr F. A., Nigg E. A. (2007) Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell 18, 4024–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eckerdt F., Maller J. L. (2008) Kicking off the polo game. Trends Biochem. Sci. 33, 511–513 [DOI] [PubMed] [Google Scholar]
- 23. Macurek L., Lindqvist A., Medema R. H. (2009) Aurora-A and hBora join the game of Polo. Cancer Res. 69, 4555–4558 [DOI] [PubMed] [Google Scholar]
- 24. Silljé H. H., Nigg E. A. (2006) Purification of mitotic spindles from cultured human cells. Methods 38, 25–28 [DOI] [PubMed] [Google Scholar]
- 25. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 26. Jensen S. S., Larsen M. R. (2007) Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun. Mass Spectrom. 21, 3635–3645 [DOI] [PubMed] [Google Scholar]
- 27. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 28. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 29. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 30. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 31. Choi H., Nesvizhskii A. I. (2008) False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J. Proteome Res. 7, 47–50 [DOI] [PubMed] [Google Scholar]
- 32. Lane H. A., Nigg E. A. (1996) Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135, 1701–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blangy A., Lane H. A., d'Hérin P., Harper M., Kress M., Nigg E. A. (1995) Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169 [DOI] [PubMed] [Google Scholar]
- 34. Elowe S., Hümmer S., Uldschmid A., Li X., Nigg E. A. (2007) Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21, 2205–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J. J., Hoffmann M., Rettig W. J., Kraut N., Peters J. M. (2007) The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17, 304–315 [DOI] [PubMed] [Google Scholar]
- 36. Matsumura S., Toyoshima F., Nishida E. (2007) Polo-like kinase 1 facilitates chromosome alignment during prometaphase through BubR1. J. Biol. Chem. 282, 15217–15227 [DOI] [PubMed] [Google Scholar]
- 37. Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. (1999) Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974 [DOI] [PubMed] [Google Scholar]
- 38. Nishizuka S., Charboneau L., Young L., Major S., Reinhold W. C., Waltham M., Kouros-Mehr H., Bussey K. J., Lee J. K., Espina V., Munson P. J., Petricoin E., 3rd, Liotta L. A., Weinstein J. N. (2003) Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc. Natl. Acad. Sci. U.S.A. 100, 14229–14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonaldi T., Straub T., Cox J., Kumar C., Becker P. B., Mann M. (2008) Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol. Cell 31, 762–772 [DOI] [PubMed] [Google Scholar]
- 40. Bose R., Molina H., Patterson A. S., Bitok J. K., Periaswamy B., Bader J. S., Pandey A., Cole P. A. (2006) Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc. Natl. Acad. Sci. U.S.A. 103, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kolkman A., Daran-Lapujade P., Fullaondo A., Olsthoorn M. M., Pronk J. T., Slijper M., Heck A. J. (2006) Proteome analysis of yeast response to various nutrient limitations. Mol. Syst. Biol. 2, 2006.0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tang L. Y., Deng N., Wang L. S., Dai J., Wang Z. L., Jiang X. S., Li S. J., Li L., Sheng Q. H., Wu D. Q., Li L., Zeng R. (2007) Quantitative phosphoproteome profiling of Wnt3a-mediated signaling network: indicating the involvement of ribonucleoside-diphosphate reductase M2 subunit phosphorylation at residue serine 20 in canonical Wnt signal transduction. Mol. Cell. Proteomics 6, 1952–1967 [DOI] [PubMed] [Google Scholar]
- 43. Goto H., Kiyono T., Tomono Y., Kawajiri A., Urano T., Furukawa K., Nigg E. A., Inagaki M. (2006) Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 8, 180–187 [DOI] [PubMed] [Google Scholar]
- 44. Qi W., Tang Z., Yu H. (2006) Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol. Biol. Cell 17, 3705–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Güttinger S., Laurell E., Kutay U. (2009) Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10, 178–191 [DOI] [PubMed] [Google Scholar]
- 46. Li H., Wang Y., Liu X. (2008) Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIalpha in cell cycle progression. J. Biol. Chem. 283, 6209–6221 [DOI] [PubMed] [Google Scholar]
- 47. Snead J. L., Sullivan M., Lowery D. M., Cohen M. S., Zhang C., Randle D. H., Taunton J., Yaffe M. B., Morgan D. O., Shokat K. M. (2007) A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem. Biol. 14, 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burkard M. E., Maciejowski J., Rodriguez-Bravo V., Repka M., Lowery D. M., Clauser K. R., Zhang C., Shokat K. M., Carr S. A., Yaffe M. B., Jallepalli P. V. (2009) Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 7, e1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J. M. (2003) Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22, 6598–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brar G. A., Kiburz B. M., Zhang Y., Kim J. E., White F., Amon A. (2006) Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature 441, 532–536 [DOI] [PubMed] [Google Scholar]
- 51. Varis A., Salmela A. L., Kallio M. J. (2006) Cenp-F (mitosin) is more than a mitotic marker. Chromosoma 115, 288–295 [DOI] [PubMed] [Google Scholar]
- 52. Zhu X., Chang K. H., He D., Mancini M. A., Brinkley W. R., Lee W. H. (1995) The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J. Biol. Chem. 270, 19545–19550 [DOI] [PubMed] [Google Scholar]
- 53. Hanisch A., Wehner A., Nigg E. A., Silljé H. H. (2006) Different Plk1 functions show distinct dependencies on Polo-box domain-mediated targeting. Mol. Biol. Cell 17, 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reindl W., Yuan J., Krämer A., Strebhardt K., Berg T. (2008) Inhibition of polo-like kinase 1 by blocking polo-box domain-dependent protein-protein interactions. Chem. Biol. 15, 459–466 [DOI] [PubMed] [Google Scholar]
- 55. Haren L., Stearns T., Lüders J. (2009) Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One 4, e5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. De Luca M., Lavia P., Guarguaglini G. (2006) A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle 5, 296–303 [DOI] [PubMed] [Google Scholar]
- 57. Chan E. H., Santamaria A., Silljé H. H., Nigg E. A. (2008) Plk1 regulates mitotic Aurora A function through betaTrCP-dependent degradation of hBora. Chromosoma 117, 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eckerdt F., Pascreau G., Phistry M., Lewellyn A. L., DePaoli-Roach A. A., Maller J. L. (2009) Phosphorylation of TPX2 by Plx1 enhances activation of Aurora A. Cell Cycle 8, 2413–2419 [DOI] [PubMed] [Google Scholar]
- 59. Ferrari S., Marin O., Pagano M. A., Meggio F., Hess D., El-Shemerly M., Krystyniak A., Pinna L. A. (2005) Aurora-A site specificity: a study with synthetic peptide substrates. Biochem. J. 390, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miller M. L., Jensen L. J., Diella F., Jørgensen C., Tinti M., Li L., Hsiung M., Parker S. A., Bordeaux J., Sicheritz-Ponten T., Olhovsky M., Pasculescu A., Alexander J., Knapp S., Blom N., Bork P., Li S., Cesareni G., Pawson T., Turk B. E., Yaffe M. B., Brunak S., Linding R. (2008) Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 1, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kufer T. A., Silljé H. H., Körner R., Gruss O. J., Meraldi P., Nigg E. A. (2002) Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bakal C., Linding R., Llense F., Heffern E., Martin-Blanco E., Pawson T., Perrimon N. (2008) Phosphorylation networks regulating JNK activity in diverse genetic backgrounds. Science 322, 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gstaiger M., Aebersold R. (2009) Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 10, 617–627 [DOI] [PubMed] [Google Scholar]
- 64. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Taylor S., Peters J. M. (2008) Polo and Aurora kinases: lessons derived from chemical biology. Curr. Opin. Cell Biol. 20, 77–84 [DOI] [PubMed] [Google Scholar]
- 66. Weiss W. A., Taylor S. S., Shokat K. M. (2007) Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat. Chem. Biol. 3, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Peters U., Cherian J., Kim J. H., Kwok B. H., Kapoor T. M. (2006) Probing cell-division phenotype space and Polo-like kinase function using small molecules. Nat. Chem. Biol. 2, 618–626 [DOI] [PubMed] [Google Scholar]
- 68. Jang C. Y., Coppinger J. A., Seki A., Yates J. R., 3rd, Fang G. (2009) Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J. Cell Sci. 122, 1334–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Blagden S. P., Glover D. M. (2003) Polar expeditions—provisioning the centrosome for mitosis. Nat. Cell Biol. 5, 505–511 [DOI] [PubMed] [Google Scholar]
- 70. Macùrek L., Lindqvist A., Lim D., Lampson M. A., Klompmaker R., Freire R., Clouin C., Taylor S. S., Yaffe M. B., Medema R. H. (2008) Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123 [DOI] [PubMed] [Google Scholar]
- 71. Seki A., Coppinger J. A., Jang C. Y., Yates J. R., Fang G. (2008) Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ahonen L. J., Kallio M. J., Daum J. R., Bolton M., Manke I. A., Yaffe M. B., Stukenberg P. T., Gorbsky G. J. (2005) Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 15, 1078–1089 [DOI] [PubMed] [Google Scholar]
- 73. Mishra R. K., Chakraborty P., Arnaoutov A., Fontoura B. M., Dasso M. (2010) The Nup107–160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12, 164–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee S. H., Sterling H., Burlingame A., McCormick F. (2008) Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22, 2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Blower M. D., Feric E., Weis K., Heald R. (2007) Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 179, 1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Montembault E., Dutertre S., Prigent C., Giet R. (2007) PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J. Cell Biol. 179, 601–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pyronnet S., Dostie J., Sonenberg N. (2001) Suppression of cap-dependent translation in mitosis. Genes Dev. 15, 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schwartz D., Gygi SP. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]