Abstract
Mass spectrometry is a fundamental tool for discovery and analysis in the life sciences. With the rapid advances in mass spectrometry technology and methods, it has become imperative to provide a standard output format for mass spectrometry data that will facilitate data sharing and analysis. Initially, the efforts to develop a standard format for mass spectrometry data resulted in multiple formats, each designed with a different underlying philosophy. To resolve the issues associated with having multiple formats, vendors, researchers, and software developers convened under the banner of the HUPO PSI to develop a single standard. The new data format incorporated many of the desirable technical attributes from the previous data formats, while adding a number of improvements, including features such as a controlled vocabulary with validation tools to ensure consistent usage of the format, improved support for selected reaction monitoring data, and immediately available implementations to facilitate rapid adoption by the community. The resulting standard data format, mzML, is a well tested open-source format for mass spectrometer output files that can be readily utilized by the community and easily adapted for incremental advances in mass spectrometry technology.
Mass spectrometry (MS)1 has recently emerged as a major discovery tool in the life sciences (1). This analytical technique is used to analyze the molecular composition of a biological sample by ionizing the sample or analyte molecules and then measuring the mass-to-charge ratios of the resulting ions. The data from an MS experiment consist of mass spectra that are used to identify, characterize, and quantify the abundance of the molecules of interest. The resulting MS spectra, along with their associated metadata (e.g. experimental protocol, MS instrumentation, operational parameters, etc.), are then semi-automatically processed by specialized software packages to identify or quantify the sampled ions. The inherent variability introduced by using different instruments, instrument software, and experimental conditions, however, affects the downstream ability to analyze, integrate, and compare data sets originating from different MS experiments.
Indeed, with the ever-increasing use of mass spectrometry, two issues have arisen in terms of handling MS data: (i) the necessity to share data throughout the scientific community in order to facilitate integration and comparison (2), and (ii) the importance of utilizing open and readily accessible standard formats that verifiably capture a consistent amount of crucial information. The importance of addressing these issues has been further emphasized in prominent journal editorials (3–4). Data repositories have since been created to allow data to be shared, including Tranche (5), GPMDB (6), PRIDE (7), and PeptideAtlas (8), among others (9), and various proposed standard formats for MS data (10–14) were developed. Other formats such as JCAMP-DX (http://www.acornnmr.com/JCAMP.htm; www.jcamp.org), which was designed for IR spectrometry and adapted to NMR and mass spectrometry, and NetCDF are quite variably implemented, difficult to validate, and cannot encode extensive metadata in a standard fashion and therefore have not gained much use for proteomics applications and other complex MS analyses. Analytical Information Markup Language (AnIML; http://animl.sourceforge.net/), which aims to encompass several analytical platforms, including eventually mass spectrometry, is still being designed. For mass spectrometry-based proteomics workflows, mzXML (13) and mzData (14) have been the most widely used open formats for several years.
However, each of these initial efforts to develop an open, vendor-neutral XML data format to store MS information was undertaken with a different underlying purpose. One format, mzData, was developed by HUPO-PSI as a data exchange and archive standard (14, 15), and was implemented as such in PRIDE (16). The other format, mzXML, was developed at the Institute for Systems Biology in an effort to streamline their data processing software (17), and became a popular de-facto standard format. These two formats also differed in their underlying philosophies regarding flexibility. mzData utilized a controlled vocabulary that could be frequently updated as the technology advanced. In contrast, mzXML had a strict schema that used enumerated attributes to describe the auxiliary information, such that support for new annotations required revisions to the schema and software updates.
Although each of the proposed formats satisfied the requirements of openness and accessibility, the multiplicity of the formats proved to be confusing and distracting to scientists and computer programmers alike. In order to resolve this situation, the teams that developed mzData and mzXML, along with many other researchers and developers from academia, industry, and vendors joined forces in the Human Proteome Organization (HUPO) Proteomics Standards Initiative working group for mass spectrometry standards (PSI-MS), and set out to create a single MS data standard that would build on the strengths of the previous efforts. The challenge in creating the new unified output format, called mzML, was therefore the resolution of the opposing philosophies of mzXML and mzData, while retaining the best technical attributes of these two formats.
History
In 2006, the unification process was initiated at a PSI workshop based on the guiding design principles determined by members representing instrument and software vendors, data repositories, end users, and the teams that built the mzXML and mzData standards. The designers of mzML focused on four key objectives: (i) creation of a simple format, (ii) elimination of alternate ways to encode the same information, (iii) support for all the features of both mzXML and mzData, and (iv) validation through implementation prior to release. Taken together, these goals would lead to a single unified format that could support the current capabilities of mzXML and mzData and that could be easily supported by vendors and current software, with further enhancements to be considered in future releases. In order to facilitate swift adoption and uniform implementation of the new standard format, the participants of PSI-MS also created open source tool sets that enabled developers as well as end users to immediately pick up the format without having to write their own software.
Progress on the format was made at regular PSI workshops as well as special workshops dedicated to mzML. In June 2008, the mzML 1.0 standard format was released (18, 19). However, despite the rather rigorous review process (20), several shortcomings became apparent as vendors quickly moved to implement the new format, most notably insufficient support for precursor ion scans and neutral loss scans, and a severe file size inflation problem for Selected Reaction Monitoring runs (all of which represented novel features that had been absent from the precursor formats). These deficiencies, along with several other minor issues, were remedied by the PSI-MS working group in collaboration with the implementers that had detected the issues. As a result, mzML version 1.1.0 was released in June 2009, with the expectation that this new version will remain stable for quite some time.
Design
In addition to incorporating the best technical attributes of the predecessor formats, several key innovations were introduced in mzML. First, in order to support new hybrid instruments such as the LTQ Orbitrap and LTQ FT, mzML can specify multiple operational configurations for an instrument, and link individual spectra to a specific configuration. Another new feature is the ability to capture Selected Reaction Monitoring data efficiently, through the newly introduced chromatogram elements. More detailed improvements are also found in mzML, such as the ability to encode isolation window size, enabling gas phase fractionation/MSe data to be correctly annotated, and accommodating the presence of multiple precursor ions within a typical liquid chromatography (LC)-MS/MS isolation window (21). Associated with mzML comes a rich, schema-linked controlled vocabulary (CV) that allows accurate and unambiguous annotation of metadata. In addition, mzML comes with a set of semantic validation rules. These rules are encoded in a mapping XML document according to the PSI Validator framework (22)(see http://www.psidev.info/validator) and have been implemented in two independent mzML validator applications (see http://www.psidev.info/index.php?q=node/390).
The full technical details of the mzML standard are available online, together with complete specification documentation, graphical depictions of its structure, and various example files at http://www.psidev.info/index.php?q=node/257. Next we will highlight the primary technical aspects of the mzML standard and discuss current implementations.
All of the information from a single MS run, including the spectra and associated metadata, is contained within the mzML file. Like its predecessors, mzML is encoded in XML. An XML schema definition defines the format structure, and many industry-standard tools are readily available to validate whether an XML document conforms to its XML schema definition.
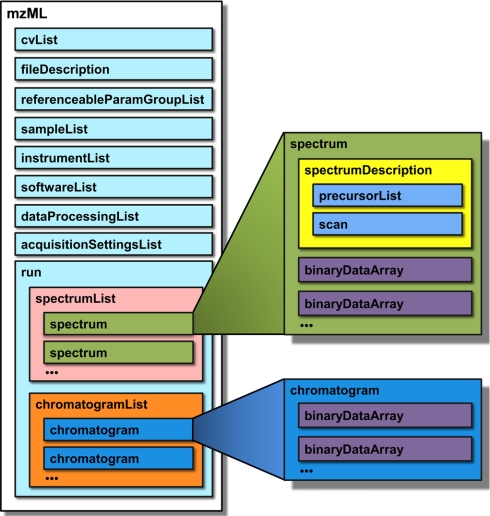
The overall mzML file structure (Fig. 1) is as follows (elements presented top-to-bottom): <cvList> contains information about the controlled vocabularies referenced in the rest of the mzML document; <fileDescription> contains basic information on the type of spectra contained in the file; <referenceableParamGroupList> is an optional element that of groups of controlled vocabulary terms that can be referenced as a unit throughout defines a list the document; <sampleList> can contain information about samples that are referenced in the file; <instrumentConfigurationList> contains information about the instrument that generated the run; <softwareList> and <dataProcessingList> provide a history of data processing that occurred after the raw acquisition; <acquisitionSettingsList> is an optional element that stores special input parameters for the mass spectrometer, such as inclusion lists. These elements are followed by the acquired spectra and chromatograms. Both spectral and chromatographic data are represented by binary format data encoded into base 64 strings, rather than human-readable ASCII text for enhanced fidelity and efficiency when dealing with profile data. This design choice does not enjoy unanimous approval, but has been agreed upon by the majority of designers.
Fig. 1.
A schematic representation of mzML, showing key elements of the format. Each rectangle represents an XML element. See main text for a full description.
In order to enable fast access to the file, mzML was designed with a standardized but optional mechanism for random access indexing, in the same way as mzXML. This enables programs to directly locate a specific spectrum within the file during processing, rather than having to read the file sequentially. Although there is debate about whether to include a random access index because of the possibility of index corruption, years of experience with mzXML have demonstrated that these problems are rare and are outweighed by the benefits of having an index. To compensate for the possibilities of an error in the index, reader software can easily be written to verify the offsets and automatically rebuild the index if there is an error. To make the index completely optional, mzML was designed so that the primary document does not have an index, but the document can still be enclosed in a wrapper schema that has an index. Thus, an mzML file may contain either a plain or indexed mzML document and reader software is designed to handle either case transparently.
Finally, although the open and standardized XML formatting provides clear advantages, it also implicitly requires a certain verbosity that enlarges the size of the data files by as much as a factor of 10 for profile-mode spectra without compression when compared with the original raw files. However, enabling in-line zlib compression typically reduces the files by a factor of 2 below the uncompressed form. Further, because any remaining size increase is primarily because of the presence of XML tags, standard and commonly used compression tools can be used to reduce this size overhead for storage and transport of mzML files. Compression factors for mzML files can vary by compression algorithm, but GZIP, zip, and 7zip (all freely available compression algorithms, supported on many platforms) provide size reductions of another factor of 2, thus essentially offsetting the size increase initially seen in uncompressed files. Profile mode spectra often undergo peak picking (if desired by the user) during conversion to mzML and therefore lead to smaller files than the original. A typical ion trap file with already centroided peaks in the original file becomes 1.8× larger with mzML without compression, or just 1.3× larger when using in-line zlib compression, and 0.5× (i.e. half as large as the original) when using both in-line compression and total file compression with the gzip algorithm. Some applications are able to work directly with gzipped mzML files, thus providing an overall savings in disk space, assuming the original files are archived elsewhere. Nonetheless, overall additional disk space costs associated with using standard formats are typically much lower than the human costs associated with trying to work with multiple proprietary formats.
Controlled Vocabulary
In an effort to prevent encoding the same information in slightly different ways and to provide support for new technologies with mzML, we have designed the format to encode most of the metadata in <cvParam> elements, which provide a reference to a specific concept within the PSI MS controlled vocabulary (CV). These CV terms have explicit and detailed definitions, including the data type and type of units required. The controlled vocabulary is adjustable and new CV terms can be added without modification of the mzML schema. Whenever an implementer requires a new term to describe a new concept, the proposed term and definition can be mailed to the PSI-MS vocabulary list (psidev-ms-vocab@lists.sourceforge.net), where the addition can be discussed and then added to the CV within hours or days. Additionally, other CVs can also be used to annotate specific elements; the NEWT ontology for species can for instance be used to annotate the sample.
Semantic Validation
To enforce the use of CV terms, a semantic validator was released with the data format. Semantic validation provides a simple yet powerful means to assess the completeness and semantic correctness of the metadata in an mzML file, automatically spotting errors such as the absence of a required binary array type annotation, the incorrect annotation of an ionization source with a detector-specific CV term, or the use of two conflicting CV terms where only one can be valid at a time. We have made the validator available as a webpage with file uploading, or as a standalone tool for local validation. Furthermore, because the mzML format is designed to support full MIAPE (10, 23) compliance, automated semantic analyses can be carried out a priori by any consumer of an mzML data file to ascertain the presence of the required minimal information. An additional benefit is that the metadata can be customized for different types of data, so that different types of spectra can be encoded using the same tags, but with different metadata.
mzML Development Process
Development of the mzML format has followed the overall HUPO PSI community standard development process, which in turn is largely based on the highly successful open source software development model. A centralized group of core volunteers takes care of coordinating the efforts of the many enthusiastic community members that contribute their time and expertise at different times, and a full record of the entire process is maintained through an online mailing list that is directly accessible to all. This development model has been proven to be (perhaps paradoxically) extremely robust as compared with more tightly organized and coordinated projects. Indeed, even though the core development team of mzML has changed substantially over the years, this never impacted the development of the standard proper.
The mzML standard is furthermore deemed quite future proof as it has been developed with change in mind. The required flexibility of the format comes primarily from its mixed structure—certain aspects of the data are rather rigidly defined in the XML format specification, such as the necessity to include an instrument description. Yet the actual form that this description takes is quite open, and not defined by the XML schema. As an example, consider the “source” element for an instrument. The different types of sources are defined solely through controlled vocabulary parameters, and if a new source is invented tomorrow, a simple update to the CV will automatically enable mzML files to communicate the use of this new source. Furthermore, because CV terms are linked through defined relationships, this new source term will be immediately recognizable to existing software as describing a source, because it will have an “is a” relationship to the metaterm “ion source.” This approach is employed in virtually every element in mzML, making the format extremely flexible without requiring any updates to either XML schema or software parsers. Changes need thus only be made to the CV, which is a simple text file that is made available in a version control system online, and that can be updated and read on-the-fly. Indeed, because the first public release of mzML, numerous updates have already been introduced to the controlled vocabulary without effecting any downstream changes on the XML schema or the existing software.
Implementations
Because of the broad community participation in PSI-MS, there are several implementations of the mzML format in software tools, legacy data converters, and programming libraries for a variety of languages (see http://www.psidev.info/index.php?q=node/257 for a current summary). In fact, the wide variety of software that uses mzML continues to grow and is one of the strengths of mzML. The ProteoWizard software project (24–25) has provided the framework for testing and reference implementation of mzML in its final stages of development. It consists of a set of open-source, cross-platform tools and libraries written in C++ for proteomic data analyses. The libraries provide a well-tested framework that unifies data file access and performs standard chemistry and LCMS dataset computations, making ProteoWizard an ideal library to include in any software project that needs to add mzML read or write support. ProteoWizard is available under a very permissive license, which allows the library to be used in commercial software without affecting the license terms of that software. The ProteoWizard library is already used by several unrelated software projects to provide mzML support. The Proteowizard “msconvert” tool can convert many different vendor formats to mzML, as well as convert mzXML files into mzML.
OpenMS (26), an open-source C++ library for mass spectrometry, also provides classes for reading and writing mzML which can be easily integrated in other software tools. Additionally, it supports both XSD validation and semantic validation of mzML files. This functionality of OpenMS was used to implement an off-line tool for validation of mzML files which is part of TOPP - The OpenMS Proteomics Pipeline (27). Similarly, the NCBI C++ toolkit and the jmzML Java toolkit (28) provide libraries for reading and writing mzML. Because these libraries are already available to simplify addition of mzML support, several software applications are already being distributed with mzML 1.1 support. These include search engines and postprocessing software such as X!Tandem (29), Myrimatch (30), the Trans-Proteomic Pipeline (TPP) (31–33), and the Proteios Software Environment (34). Most vendors have committed to provide mzML support in the next release of their software.
The widespread support for mzML in existing, commercial tools, along with the availability of several production-grade open source software packages and libraries in a variety of programming languages, ensures that data encoded in the mzML format is readily accessible to any interested end user or software developer.
Example Usages
Because the main advantages of open data standards over closed, proprietary formats are interoperability and portability, we have chosen two corresponding use cases in the field of mass spectrometry-based proteomics to illustrate some of the usages of the mzML data standard. First, many laboratories employ multiple instruments from different vendors for their analyses. Although this heterogeneity in instrumentation confers the important advantage of providing complementary strengths of the different machines, it also creates a logistical problem at the level of data processing. The various proprietary data formats employed by each instrument to report its data, are essentially tied to these specific instruments - even different models from the same vendor can deliver incompatible output files. As a result, the development of software that can operate on data from any instrument, such as the tools in the Trans-Proteomic Pipeline, becomes quite difficult indeed. This in fact was one of the main reasons why the original mzXML format was developed as part of the Trans-Proteomic Pipeline: to unify the various vendor formats in a common, open data structure that maintains sufficient amounts of data to reliably support various kinds of downstream processing, including identification and quantification of proteins. As a direct descendant from mzXML, mzML provides these same benefits, allowing data from many instruments to be transformed (using the freely available ProteoWizard or TPP tools) into the common mzML format, which is in turn read and interpreted homogenously by all downstream data processing software applications.
A second important use case of standard data formats concerns the dissemination of data to the wider scientific community, an endeavor that is very deeply ingrained in the life sciences (35). If data were disseminated in proprietary formats, three problems would occur (discussed in detail in (36)): (i) referees wishing to evaluate (privately) deposited data during peer review would have difficulties accessing, interpreting, and validating the data and derived conclusions unless they happened to own the same instruments and software compatible with the format, (ii) after publication of the data, interested consumers would face similar difficulties in accessing and processing the data, and (iii) over a relatively short time span, all data would become unreadable, as the required vendor-specific software will no longer be supported or available. By employing an open, XML (and therefore ultimately text-based) format such as mzML, these three key issues are implicitly circumvented.
Both of these examples, of course, rely on the availability of software supporting the format, but as can be seen from the previous section, many actively supported free and open source implementations in a variety of programming languages and for a variety of platforms are already available for mzML today, and many other implementations are underway or will be available with their next software release. Finally, it should be noted that the two use cases are in fact connected: by switching to mzML as the format for within-lab data processing and analysis, the step to disseminate in mzML becomes effectively trivial.
Integration with Other Standards in the Life Sciences
The data accommodated by mzML will most likely not stand alone in a modern-day workflow. Preceded by sample treatment and sample separation (often through chromatography), mass spectrometry data is then usually further processed to identify or quantify the recorded signals. As such, it is important to note that HUPO PSI has also released standards for protein separation including gel based and column chromatography based methods (http://www.psidev.info/index.php?q=node/83), for identification of molecules from mass spectra (http://www.psidev.info/index.php?q=node/319), and for the annotation of modifications on proteins (http://www.psidev.info/index.php?q=node/319). Furthermore, the overall integration of standardized data and metadata across domains in the life sciences is being actively undertaken by the Reporting Structure for Biological Investigations (RSBI) working group of the MGED Society (http://www.mged.org), which has culminated in the ISA-TAB format (37). Minimal information assurance in all the relevant formats on the other hand is coordinated through the MIBBI project (38).
CONCLUSION
In 2009, three years after its conception, mzML 1.1 was released and has proven to be a solid format that can easily accommodate incremental advances in mass spectrometry technology, while providing a good foundation for extension to accommodate encoding of data from new technologies. An existing set of software libraries that support mzML will enable quick adoption of the format. However, because the precursor formats are also highly capable, the incentive to migrate existing workflows is low, and the adoption of mzML in practice will be gradual. An initial wave of implementations necessitated a revision of 1.0 to 1.1, but since the release of 1.1, there have not been any significant changes necessary. It is therefore expected that 1.1 will remain stable for quite some time. The involvement of instrument vendors in PSI-MS further ensures that mzML export will become available on instrument software by default.
Like all PSI standards, mzML 1.1 has gone through a formal review process called the PSI document process (20), which consists of three review periods managed by the PSI Editor: an internal review, an external review by invited experts, and a public review stage. As such, we believe that mzML 1.1 can now readily be utilized by the community at large, providing a single, open, and accessible community standard format for mass spectrometer output files. With CV annotations, semantic validation, and MIAPE compliance as part of the design of the standard, unambiguous reporting of metadata will thus become standard practice, ensuring that mzML can be used as a highly reliable data exchange format. The PSI-MS working group will meanwhile continue to refine the controlled vocabulary and coordinate software development surrounding mzML to ensure that mzML stays up-to-date with the progress of the field.
Acknowledgments
We would like to thank all individuals who have contributed in PSI meetings, mailing lists, and elsewhere with ideas, comments, and implementation efforts on mzML, especially Ruedi Aebersold, Rolf Apweiler, Ron Beavis, Benito Cañas, Mike Coleman, David Creasy, Eva Duchoslav, Jimmy Eng, Jayson Falkner, Lola Gutierrez, Jari Häkkinen, David Horn, Phil Jones, Marius Kallhardt, Jim Langridge, Kent Laursen, Parag Mallick, Ruth McNally, Alberto Medina, Luis Mendoza, Lars Nilse, Erik Nilsson, Sandra Orchard, Norman Paton, Patrick Pedrioli, Rune Philosof, Brian Pratt, Howard Read, Sean Seymour, David Shteynberg, David Sparkman, Chris Taylor, and Trish Wheztel. We thank Julie Bletz, Terry Farrah, and Gordon Sun for assistance with preparation of the manuscript. L.M. and F.R. would like to thank Rolf Apweiler for his support, and L.M. would additionally like to thank Joël Vandekerckhove for support.
Footnotes
* L.M. is supported by the “ProDaC” grant LSHG-CT-2006-036814 of the European Union, F.R. by the Wellcome Trust [grant number WT085949MA], and E.W.D. by the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract No. N01-HV-28179 and by the Duchy of Luxembourg.
We declare the following competing financial or commercial interests:
Several authors work for commercial entities whose software supports the mzML format.
1 The abbreviations used are:
- MS
- mass spectrometry
- HUPO
- Human Proteome Organization
- PSI-MS
- Proteomics Standards Initiative working group for mass spectrometry standards
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- CV
- controlled vocabulary.
REFERENCES
- 1. Editors (2007) Mind the technology gap. Nat. Methods 4, 765. [DOI] [PubMed] [Google Scholar]
- 2. Prince J. T., Carlson M. W., Wang R., Lu P., Marcotte E. M. (2004) The need for a public proteomics repository. Nature Biotechnology 22, 471–472 [DOI] [PubMed] [Google Scholar]
- 3. Editors, (2008) Thou shalt share your data. Nat. Methods 5, 209 [Google Scholar]
- 4. Editors, (2007) Democratizing proteomics data. Nat Biotechnol 25, 262. [DOI] [PubMed] [Google Scholar]
- 5. Falkner J. A., Andrews P. C. (2007) Tranche: Secure Decentralized Data Storage for the proteomics community. Journal of Biomolecular Techniques 18, 3 [Google Scholar]
- 6. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 7. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. (2005) PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 8. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res 34, D655–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mead J. A., Bianco L., Bessant C. (2009) Recent developments in public proteomic MS repositories and pipelines. Proteomics 9, 861–881 [DOI] [PubMed] [Google Scholar]
- 10. Taylor C. F., Binz P. A., Aebersold R., Affolter M., Barkovich R., Deutsch E. W., Horn D. M., Huhmer A., Kussmann M., Lilley K., Macht M., Mann M., Muller D., Neubert T. A., Nickson J., Patterson S. D., Raso R., Resing K., Seymour S. L., Tsugita A., Xenarios I., Zeng R., Julian R. K., Jr. (2008) Guidelines for reporting the use of mass spectrometry in proteomics. Nat Biotechnol. 26, 860–861 [DOI] [PubMed] [Google Scholar]
- 11. McDonald W. H., Tabb D. L., Sadygov R. G., MacCoss M. J., Venable J., Graumann J., Johnson J. R., Cociorva D., Yates J. R., 3rd (2004) MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 12. Orchard S., Montechi-Palazzi L., Deutsch E. W., Binz P. A., Jones A. R., Paton N., Pizarro A., Creasy D. M., Wojcik J., Hermjakob H. (2007) Five years of progress in the Standardization of Proteomics Data 4(th) Annual Spring Workshop of the HUPO-Proteomics Standards Initiative April 23–25, 2007 Ecole Nationale Superieure (ENS), Lyon, France. Proteomics 7, 3436–3440 [DOI] [PubMed] [Google Scholar]
- 13. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 14. mzData, http://psidev.info/index.php?q=node/80#mzdata.
- 15. Orchard S., Zhu W., Julian R. K., Jr., Hermjakob H., Apweiler R. (2003) Further advances in the development of a data interchange standard for proteomics data. Proteomics 3, 2065–2066 [DOI] [PubMed] [Google Scholar]
- 16. Jones P., Cote R. G., Martens L., Quinn A. F., Taylor C. F., Derache W., Hermjakob H., Apweiler R. (2006) PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res 34, D659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deutsch E. (2008) mzML: a single, unifying data format for mass spectrometer output. Proteomics 8, 2776–2777 [DOI] [PubMed] [Google Scholar]
- 19. Deutsch E. W. (2010) Mass spectrometer output file format mzML. Methods Mol. Biol. 604, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vizcaino J. A., Martens L., Hermjakob H., Julian R. K., Paton N. W. (2007) The PSI formal document process and its implementation on the PSI website. Proteomics 7, 2355–2357 [DOI] [PubMed] [Google Scholar]
- 21. Luethy R., Kessner D. E., Katz J. E., Maclean B., Grothe R., Kani K., Faca V., Pitteri S., Hanash S., Agus D. B., Mallick P. (2008) Precursor-ion mass re-estimation improves peptide identification on hybrid instruments. J Proteome Res 7, 4031–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Montecchi-Palazzi L., Kerrien S., Reisinger F., Aranda B., Jones A. R., Martens L., Hermjakob H. (2009) The PSI semantic validator: a framework to check MIAPE compliance of proteomics data. Proteomics 9, 5112–5119 [DOI] [PubMed] [Google Scholar]
- 23. Taylor C. F., Paton N. W., Lilley K. S., Binz P. A., Julian P. K., Jr., Jones A. R., Zhu W., Apweiler R., Aebersold R., Deutsch E. W., Dunn M. J., Heck A. J., Leitner A., Macht M., Mann M., Martens L., Neubert T. A., Patterson S. D., Ping P., Seymour S. L., Souda P., Tsugita A., Vandekerckhove J., Vondriska T. M., Whitelegge J. P., Wilkins M. R., Xenarios I., Yates J. R., 3rd, Hermjakob H. (2007) The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol. 25, 887–893 [DOI] [PubMed] [Google Scholar]
- 24. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. ProteoWizard, http://proteowizard.sourceforge.net
- 26. Sturm M., Bertsch A., Gropl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., Kohlbacher O. (2008) OpenMS-An open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kohlbacher O., Reinert K., Gropl C., Lange E., Pfeifer N., Schulz-Trieglaff O., Sturm M. (2007) TOPP-The OpenMS proteomics pipeline. Bioinformatics 23, e191–197 [DOI] [PubMed] [Google Scholar]
- 28. Cote R. G., Reisinger F., Martens L. (2010) jmzML, an open-source Java API for mzML, the PSI standard for MS data. Proteomics 10, 1332–1335 [DOI] [PubMed] [Google Scholar]
- 29. Bjornson R. D., Carriero N. J., Colangelo C., Shifman M., Cheung K. H., Miller P. L., Williams K. (2008) X!!Tandem, an improved method for running X!tandem in parallel on collections of commodity computers. J Proteome Res 7, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pedrioli P. G. (2010) Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol Biol 604, 213–238 [DOI] [PubMed] [Google Scholar]
- 33. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hakkinen J., Vincic G., Mansson O., Warell K., Levander F. (2009) The proteios software environment: an extensible multiuser platform for management and analysis of proteomics data. J Proteome Res 8, 3037–3043 [DOI] [PubMed] [Google Scholar]
- 35. Vizcaíno J. A., Mueller M., Hermjakob H., Martnes L. (2009) Charting online OMICS resources: a navigational chart for clinical researchers. Proteomics Clinical Applications 3, 18–29 [DOI] [PubMed] [Google Scholar]
- 36. Martens L., Nesvizhskii A. I., Hermjakob H., Adamski M., Omenn G. S., Vandekerckhove J., Gevaert K. (2005) Do we want our data raw? Including binary mass spectrometry data in public proteomics data repositories. Proteomics 5, 3501–3505 [DOI] [PubMed] [Google Scholar]
- 37. Sansone S. A., Rocca-Serra P., Brandizi M., Brazma A., Field D., Fostel J., Garrow A. G., Gilbert J., Goodsaid F., Hardy N., Jones P., Lister A., Miller M., Morrison N., Rayner T., Sklyar N., Taylor C., Tong W., Warner G., Wiemann S. (2008) The first RSBI (ISA-TAB) workshop: “can a simple format work for complex studies?”. OMICS 12, 143–149 [DOI] [PubMed] [Google Scholar]
- 38. Taylor C. F., Field D., Sansone S. A., Aerts J., Apweiler R., Ashburner M., Ball C. A., Binz P. A., Bogue M., Booth T., Brazma A., Brinkman R. R., Michael Clark A., Deutsch E. W., Fiehn O., Fostel J., Ghazal P., Gibson F., Gray T., Grimes G., Hancock J. M., Hardy N. W., Hermjakob H., Julian R. K., Jr., Kane M., Kettner C., Kinsinger C., Kolker E., Kuiper M., Novere N. L., Leebens-Mack J., Lewis S. E., Lord P., Mallon A. M., Marthandan N., Masuya H., McNally R., Mehrle A., Morrison N., Orchard S., Quackenbush J., Reecy J. M., Robertson D. G., Rocca-Serra P., Rodriguez H., Rosenfelder H., Santoyo-Lopez J., Scheuermann R. H., Schober D., Smith B., Snape J., Stoeckert C. J., Jr., Tipton K., Sterk P., Untergasser A., Vandesompele J., Wiemann S. (2008) Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol 26, 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]