Abstract
Glycosylation is one of the most important and common forms of protein post-translational modification that is involved in many physiological functions and biological pathways. Altered glycosylation has been associated with a variety of diseases, including cancer, inflammatory and degenerative diseases. Glycoproteins are becoming important targets for the development of biomarkers for disease diagnosis, prognosis, and therapeutic response to drugs. The emerging technology of glycoproteomics, which focuses on glycoproteome analysis, is increasingly becoming an important tool for biomarker discovery. An in-depth, comprehensive identification of aberrant glycoproteins, and further, quantitative detection of specific glycosylation abnormalities in a complex environment require a concerted approach drawing from a variety of techniques. This report provides an overview of the recent advances in mass spectrometry based glycoproteomic methods and technology, in the context of biomarker discovery and clinical application.
With recent advances in proteomics, analytical and computational technologies, glycoproteomics—the global analysis of glycoproteins—is rapidly emerging as a subfield of proteomics with high biological and clinical relevance. Glycoproteomics integrates glycoprotein enrichment and proteomics technologies to support the systematic identification and quantification of glycoproteins in a complex sample. The recent development of these techniques has stimulated great interest in applying the technology in clinical translational studies, in particular, protein biomarker research.
While glycomics is the study of glycome (repertoire of glycans), glycoproteomics focuses on studying the profile of glycosylated proteins, i.e. the glycoproteome, in a biological system. Considerable work has been done to characterize the sequences and primary structure of the glycan moieties attached to proteins (1–3), and their structural alterations related to cancer (4–6). Recent reports have provided a comprehensive overview of the concept of glycomics and its prospective in biomarker research (7–10). In contrast, this review is focused on recent developments in glycoproteomic techniques and their unique application and technical challenge to biomarker discovery.
Glycoproteomics in Biomarker Discovery and Clinical Study
Most secretory and membrane-bound proteins produced by mammalian cells contain covalently linked glycans with diverse structures (2). The glycosylation form of a glycoprotein is highly specific at each glycosylation site and generally stable for a given cell type and physiological state. However, the glycosylation form of a protein can be altered significantly because of changes in cellular pathways and processes resulting from diseases, such as cancer, inflammation, and neurodegeneration. Such disease-associated alterations in glycoproteins can happen in one or both of two ways: 1) protein glycosylation sites are either hypo, hyper, or newly glycosylated and/or; 2) the glycosylation form of the attached carbohydrate moiety is altered. In fact, altered glycosylation patterns have long been recognized as hallmarks in cancer progression, in which tumor-specific glycoproteins are actively involved in neoplastic progression and metastasis (5, 6, 11, 12). Sensitive detection of such disease-associated glycosylation changes and abnormalities can provide a unique avenue to develop glycoprotein biomarkers for diagnosis and prognosis. In addition, intervention in the glycosylation and carbohydrate-dependent cellular pathways represent a potential new modality for cancer therapies (6, 11, 13). Table I lists some of the FDA approved cancer biomarkers (14, 15) that are glycosylated proteins or protein complexes.
Table I. Listing of some of the US Food and Drug Administration (FDA) approved cancer biomarkers.
| Protein target | Glycosylation | Detection | Source | Disease | Clinical biomarker |
|---|---|---|---|---|---|
| α-Fetoprotein | Yes | Glycoprotein | Serum | Nonseminomatous testicular cancer | Diagnosis |
| Human chorionic gonadotropin-β | Yes | Glycoprotein | Serum | Testicular cancer | Diagnosis |
| CA19–9 | Yes | Carbohydrate | Serum | Pancreatic cancer | Monitoring |
| CA125 | Yes | Glycoprotein | Serum | Ovarian cancer | Monitoring |
| CEA (carcinoembryonic antigen) | Yes | Protein | Serum | Colon cancer | Monitoring |
| Epidermal growth factor receptor | Yes | Protein | Tissue | Colon cancer | Therapy selection |
| KIT | Yes | Protein (IHC) | Tissue | Gastrointestinal (GIST) cancer | Diagnosis/Therapy selection |
| Thyroglobulin | Yes | Protein | Serum | Thyroid cancer | Monitoring |
| PSA-prostate-specific antigen (Kallikrein 3) | Yes | Protein | Serum | Prostate cancer | Screening/Monitoring/Diagnosis |
| CA15–3 | Yes | Glycoprotein | Serum | Breast cancer | Monitoring |
| CA27–29 | Yes | Glycoprotein | Serum | Breast cancer | Monitoring |
| HER2/NEU | Yes | Protein (IHC), Protein | Tissue, Serum | Breast cancer | Prognosis/Therapy selection/Monitoring |
| Fibrin/FDP-fibrin degradation protein | Yes | Protein | Urine | Bladder cancer | Monitoring |
| BTA-bladder tumour-associated antigen (Complement factor H related protein) | Yes | Protein | Urine | Bladder cancer | Monitoring |
| CEA and mucin (high molecular weight) | Yes | Protein (Immunofluorescence) | Urine | Bladder cancer | Monitoring |
Protein biomarker development is a complex and challenging task. The criteria and approach applied for developing each individual biomarker can vary, depending on the purpose of the biomarker and the performance requirement for its clinical application (16, 17). In general, it has been suggested that the preclinical exploratory phase of protein biomarker development can be technically defined into four stages (18), including initial discovery of differential proteins; testing and selection of qualified candidates; verification of a subset of candidates; assay development and pre-clinical validation of potential biomarkers. Thanks to recent technological advances, mass spectrometry based glycoproteomics is now playing a major role in the initial phase of discovering aberrant glycoproteins associated with a disease. Glycoprotein enrichment techniques, coupled with multidimensional chromatographic separation and high-resolution mass spectrometry have greatly enhanced the analytical dynamic range and limit of detection for glycoprotein profiling in complex samples such as plasma, serum, other bodily fluids, or tissue. In addition, candidate-based quantitative glycoproteomics platforms have been introduced recently, allowing targeted detection of glycoprotein candidates in complex samples in a multiplexed fashion, providing a complementary tool for glycoprotein biomarker verification in addition to antibody based approaches. It is clear that glycoproteomics is gaining momentum in biomarker research.
Glycoproteomics Approaches
Glycoproteomic analysis is complicated not only by the variety of carbohydrates, but also by the complex linkage of the glycan to the protein. Glycosylation can occur at several different amino acid residues in the protein sequence. The most common and widely studied forms are N-linked and O-linked glycosylation. O-linked glycans are linked to the hydroxyl group on serine or threonine residues. N-linked glycans are attached to the amide group of asparagine residues in a consensus Asn-X-Ser/Thr sequence (X can be any amino acid except proline) (19). Other known, but less well studied forms of glycosylation include glycosylphosphatidylinositol anchors attached to protein carboxyl terminus, C-glycosylation that occurs on tryptophan residues (20), and S-linked glycosylation through a sulfur atom on cysteine or methionine (21, 22). Our following discussion is focused on glycoproteomic analysis of the most common N-linked and O-linked glycoproteins.
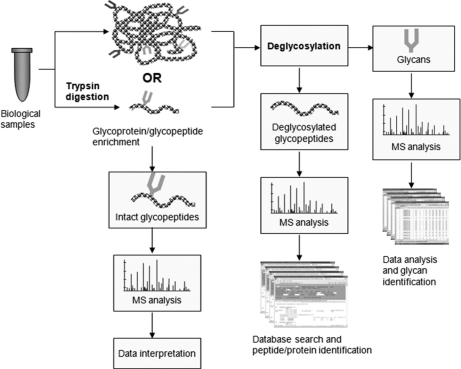
A comprehensive analysis of glycoproteins in a complex biological sample requires a concerted approach. Although the specific methods for sample preparation can be different for different types of samples (e.g. plasma, serum, tissue, and cell lysate), a glycoproteomics pipeline typically consists of glycoprotein or glycopeptide enrichment, multidimensional protein or peptide separation, tandem mass spectrometric analysis, and bioinformatic data interpretation. For glycoprotein-based enrichment methods, proteolytic digestion can be performed before or after glycan cleavage, depending on the specific workflow and enrichment methods used. For glycopeptide enrichment, proteolytic digestion is typically performed before the isolation step so that glycopeptides, instead of glycoproteins, can be captured. For quantitative glycoproteomics profiling, additional steps, such as differential stable isotope labeling of the sample and controls, are required. Fig. 1 illustrates the general strategy for an integrated glycoproteomics analysis.
Fig. 1.
The strategies of mass spectrometry based glycoproteomic analysis.
Glycoproteins or glycopeptides can be effectively enriched using a variety of techniques (see below). Following the enrichment step, the workflow then splits into two directions: glycan analysis and glycoprotein analysis. The strategies for glycan analysis have been discussed in several reviews and will not be covered in this report. For glycoprotein analysis, bottom-up workflows (“shotgun proteomics”—peptide based proteomics analysis) (23) are still most common, providing not only detailed information of a glycoprotein profile, but also the specific mapping of glycosylation sites. It is notable that the reliable analysis of mass spectrometric data in glycoproteomic studies largely relies on bioinformatic tools and glyco-related databases that are available. An increasing number of algorithms and databases for glycan analysis have been developed and well documented in several recent reviews (24–26). For glycoprotein and glycopeptide sequence analysis, a large number of well-characterized and annotated glycoproteins can be found in the UniProt Knowledgebase. In addition, many glycopeptide mass spectra are now available in the continually expanding PeptideAtlas library (27), which stores millions of high-resolution peptide fragment ion mass spectra acquired from a variety of biological and clinical samples for peptide and protein identification. Ultimately, all the data obtained from different aspects of the workflow need to be merged and interpreted in an integrated fashion so that the full extent of glycosylation changes associated with a particular biological state can be better revealed. To the best of our knowledge, the complete glycoform analysis of any glycoprotein in a specific cell type under any specific condition has not yet been accomplished for any glycoprotein with multiple glycosylation sites. Current technology can define the glycan compliment and profile the glycoproteins, but is not capable of putting them together to define the molecular species present. To date, such integrated studies still remain highly challenging, even with advanced tandem mass spectrometry technologies and growing bioinformatic resources (26, 28–31).
Enrichment of the Glycoproteome
Characterization of the glycoproteome in a complex biological sample such as plasma, serum, or tissue, is analytically challenging because of the enormous complexity of protein and glycan constituents and the vast dynamic range of protein concentration in the sample. The selective enrichment of the glycoproteome is one of the most efficient ways to simplify the enormous complexity of a biological sample to achieve an in-depth glycoprotein analysis. Two approaches for glycoprotein enrichment have been widely applied: lectin affinity based enrichment methods (31–36) and hydrazide chemistry-based solid phase extraction methods (37–42). Recent studies have demonstrated that the two methods are complementary and a very effective means for the enrichment of glycoproteins or glycopeptides from human plasma and other bodily fluids (38, 39, 43). In addition, glycoprotein and glycopeptide enrichment using boronic acid (44, 45), size-exclusion chromatography (46), hydrophilic interaction (47) and a graphite powder microcolumn (48) have been reported.
Lectin affinity enrichment is based on the specific binding interaction between a lectin and a distinct glycan structure attached on a glycoprotein (49, 50). There are a variety of lectin species that can selectively bind to different oligosaccharide epitopes. For instance, concanavalin A (ConA) binds to mannosyl and glucosyl residues of glycoproteins (51); wheat germ agglutinin (WGA) binds to N-acetyl-glucosamine and sialic acid (52); and jacalin (JAC) specifically recognizes galactosyl (β-1,3) N acetylgalactosamine and O-linked glycoproteins (53). Lectin affinity enrichment has been designed to enrich glycoproteins with specific glycan attachment from plasma, serum, tissue, and other biological samples through affinity chromatography and other methods. Multiple lectin species can also be combined to isolate multiple types of glycoproteins in complex biological samples (54–59). Concanavalin A and wheat germ agglutinin, as well as jacalin are often used together to achieve a more extensive glycproteome characterization (31, 34, 57, 59, 60). Several reports have demonstrated a multilectin column approach to achieve a global enrichment of glycoproteins with various glycan attachments from serum and plasma (31, 34, 59, 61, 62). A recent study has developed a “filter aided sample preparation (FASP)” based method, which allows highly efficient enrichment of glycopeptides using multi-lectins (63). To date, most of the work using lectin affinity for targeted glycoprotein enrichment has focused on N-glycosylation because the binding specificity of lectin for O-glycosylation is less satisfactory. To overcome such caveat, efforts have been made using serial lectin columns of concanavalin A and jacelin in tandem to isolate O-glycopeptides from human serum (35).
A hydrazide chemistry-based method has been applied to isolate glycoproteins and glycopeptides through the formation of covalent bonding between the glycans and the hydrazide groups (37). The carbohydrates on glycoproteins are first oxidized to form aldehyde groups, which sequentially react with hydrazide groups that are immobilized on a solid surface. The chemical reaction conjugates the glycoproteins to the solid phase by forming the covalent hydrazone bond. Although, conceptually, the majority of the glycoproteins in a biological sample can be captured using this method, the further analysis of the captured glycoproteins is practically limited by the method that can cleave glycoproteins or glycopeptides from the solid phase. Because there is a lack of efficient enzymes or chemicals that can specifically deglycosylate and/or release O-linked glycoproteins or glycopeptides from the solid phase, most of the studies have applied this method solely for N-linked glycoprotein analysis. PNGase F is the enzyme that can specifically release an N-glycosylated proteins or peptides (except those carrying α1→3 linked core fucose (38)) from its corresponding oligosaccharide groups. The hydrazide chemistry method is not only highly efficient in enriching N-linked glycoproteins or glycopeptides from a complex environment, but also allows great flexibility in its applications, such as capturing extracellular N-glycoproteins on live cells to monitor their abundant changes because of cell activation, differentiation, or other cellular activities (64). This method can be readily automated for analyzing a large quantity of samples.
Recent studies have compared glycoprotein isolation methods. One study assessed lectin-based protocols and hydrophilic interaction chromatography for their performance in enriching glycoproteins and glycopeptides from serum (65). Other studies compared lectin affinity and hydrazide chemistry methods for their efficiency in isolating glycoproteins and glycopeptides from a complex biological sample (39, 66, 67). The methods are complementary in enriching glycoproteins because of their different mechanisms of glycoprotein capturing. When both methods were applied, it significantly improves the coverage of the glycoproteome, resulting in an increased number of glycoproteins identified. The lectin affinity method can be tailored to target glycoproteins with specific glycan structure(s) for isolation using different lectins, thus, affording flexibility for its application in glycoproteomic studies. The application of hydrazide chemistry method has been widely used for N-linked glycosylation study. The hydrazide chemistry essentially reacts with all the proteins with carbonyl groups, which may include glycoproteins with oxidized glycans (37, 40) and other oxidized proteins that carry carbonyl groups (68–70). The high specificity of this method may mainly result from the specificity of PNGase F, the enzyme cleaving N-glycosidic bonds to release N-glycoproteins and peptides from the solid phase. This method affords high efficiency and specificity in enriching N-linked glycoproteins or glycopeptides from a complex sample, and can be easily incorporated into a proteomics workflow for integrated analysis. In addition to the lectin and hydrazide chemistry-based methods, it has been suggested that boronic acid-based solid phase extraction may also be useful for an overall glycoproteome enrichment (44, 45), on the basis of the evidence that boronic acid can form diester bonds with most glycans, including both N-linked and O-linked glycosylation (71).
Mass Spectrometric Analysis of Glycoproteome
Mass spectrometry, because of its high sensitivity and selectivity, has been one of the most versatile and powerful tools in glycoprotein analysis, to identify the glycoproteins, evaluate glycosylation sites, and elucidate the oligosaccharide structures (56, 72, 73). The utility of a top-down approach (intact protein based proteomics analysis) (74) for glycoprotein characterization in a complex sample is still technically challenging with the current technology. The most versatile and widely used current glycoproteomics methods are based on characterizing glycopeptides generated by the digestion of glycoproteins, analyzing either deglycosylated glycopeptides or intact glycopeptides with glycan attachment, as illustrated in Fig. 1.
The direct analysis of intact glycopeptides with carbohydrate attachments is complicated by the mixed information obtained from the fragment ion spectra, which may include fragment ions from the peptide backbone, the carbohydrate group and the combinations of both. Although it is technically challenging to comprehensively analyze intact glycopeptides in a global scale for a complex biological sample, complementary information regarding peptide backbone and glycan structure can likely be obtained in a single measurement. Early work using collision-induced dissociation (CID)1 has identified a few key features that are characteristics of the fragmentation of glycopeptides, providing the basis for intact glycopeptide identification (75–79). The analysis of intact glycopeptides has been carried out using a variety of different instruments, including electrospray ionization (EST)-based ion trap (IT) (80–84), quadrupole ion trap (QIT) (85–87), Fourier transform ion cyclotron resonance (FTICR) (31, 57, 88, 89), ion trap/time-of-flight (IT/TOF) (90, 91), and quadrupole/time-of-flight (Q/TOF) (92–97); matrix-assisted laser desorption/ionization (MALDI) based Q/TOF (98–100), quadrupole ion trap/time-of-flight (QIT/TOF) (86, 101, 102), and tandem time-of-flight (TOF/TOF) (81, 82, 101, 103–105) mass spectrometers. In general, the CID generated MS/MS spectrum of a glycopeptide is dominated by B- and Y-type glycosidic cleavage ions (carbohydrate fragments) (106), and b- and y-type peptide fragments from the peptide backbone. However, the MS/MS fragmentation data obtained from different instruments can have pronounced difference in providing structure information on glycan and peptide backbone, depending on the experimental setting and instrumentation used for mass analysis, including ionization methods, collision techniques and mass analyzers. Low energy CID with electrospray ionization-based ion trap, Fourier transform-ion cyclotron resonance, and Q/TOF instrument predominantly generates fragments of glycosidic bonds. The increase of collision energy using Fourier transform-ion cyclotron resonance, and Q/TOF instruments result in the more efficient fragmentation of b- and y- ions from the peptide backbone. MALDI ionization generates predominantly singly charged precursor ions, which are more stable and usually fragmented using higher energies via CID or post-source decay (PSD), generating fragments from both the peptide backbone and the glycan (98–100, 103, 107–110). Although Q/TOF instruments have been widely used for intact glycopeptide characterization, one unique feature of the ion trap instrument is that it allows repeated ion isolation/CID fragmentation cycles, which can provide a wealth of complementary information to interpret the structure of a glycan moiety and peptide backbone (56, 86, 111). Recently, fragmentation techniques using different mechanisms from CID have been introduced and applied for glycopeptide analysis, including infrared multiphoton dissociation (IRMPD) (112–115), electon-capture dissociation (ECD) (112–120) and electron-transfer disassociation (ETD) (85, 121–123). The application of infrared multiphoton dissociation and electon-capture dissociation is largely performed with Fourier transform-ion cyclotron resonance instruments. Complementary to CID fragmentation, electon-capture dissociation and electron-transfer disassociation tend to cleave the peptide backbone with no loss of the glycan moiety, providing specific information on localizing the glycosidic modification. More details regarding mass spectrometric analysis of intact glycopeptides can be found in recent reviews (56, 124). Although great efforts have been made to apply a variety of mass spectrometry techniques to study both N-linked (32, 56, 86, 87, 112–114, 125–130) and O-linked (90, 116, 119, 120, 130–140) glycopeptides, the interpretation of the fragment spectrum of an intact glycopeptide still requires intensive manual assignment and evaluation. A recent study has demonstrated the feasibility to develop an automated workflow for analyzing intact glycopeptides in mixtures (141). In general, however, a high throughput, large scale profiling of intact glycopeptides in a complex sample still remains a challenge with current technology.
The analysis of deglycosylated peptides requires the removal of glycan attachments from glycopeptides. Fortunately, for N-linked glycopeptides, the N-glycosidic bond can be specifically cleaved using the enzyme PNGase F, providing deglycosylated peptides, which can then be analyzed directly using shotgun proteomics. The PNGase F-catalyzed deglycosylation results in the conversion of asparagine to aspartic acid in the glycopeptide sequence, which introduces a mass difference of 0.9840 Da. Such distinct mass differences can be used to precisely map the N-linked glycosylation sites using high resolution mass spectrometers. Stable isotope labeling introduced by enzymatic cleavage of glycans in H218O has also been used to enhance the precise identification of N-glycosylation sites (33, 142, 143). The removal of O-linked glycans is less straightforward, most assays rely on chemical deglycosylation methods, such as trifluoromethansulfonic acid (144), hydrazinolysis (145), β-elimination (146), and periodate oxidation (35, 147). The application of these methods suffers from a variety of limitations, such as low specificity for O-linked glycosylation, degradation of the peptide backbone, and modifications of the amino acid residues—all of which can complicate or compromise O-linked glycoproteomics analysis in a complex sample. Most of the large scale glycoproteomics studies using the deglycosylation approach have been focused on N-glycoproteins, which are prevalent in blood and a rich source for biomarker discovery. O-glycosylation lacks a common core, consensus sequence, and universal enzyme that can specifically remove the glycans from the peptide backbone, thus, is more challenging to analyze for large scale profiling.
Following deglycosylation, the glycopeptides can be treated and analyzed as stripped peptides using a shotgun proteomics pipeline. MS/MS fragment spectra with b-ions and y-ions generated from CID are searched against protein databases using search algorithms, such as SEQUEST (148), MASCOT (149), and X!tandem (150), and subsequently validated via statistical analysis (151–154), to provide peptide and protein identifications with known false discovery rate. The N-glycosylation sites can be precisely mapped using the consensus sequence of Asn-X-Ser/Thr, in which asparagine is converted to aspartic acid following enzyme cleavage introducing a mass difference of 0.9840 Dalton. A variety of mass spectrometers have been used to analyze glycoproteins, in particular N-linked glycoproteins, in complex biological and clinical samples using the deglycosylation approach. These studies include electrospray ionization-based ion trap (37–39, 41, 67, 155–157), Orbitrap (158), Q/TOF (33, 35, 142, 155), triple quadrupole (159), Fourier transform-ion cyclotron resonance (64, 160); and MALDI based TOF/TOF (41, 161) and Q/TOF (37). Recently, an attempt was made to apply ion mobility-mass spectrometry (IM-MS) to characterize deglycosylated glycopeptides and the corresponding carbohydrates simultaneously (162) in a single measurement. The approach of analyzing deglycosylated glycopeptides makes it possible to utilize available proteomics technology for large-scale glycoproteome profiling, especially N-linked glycoproteins, in a high-throughput fashion.
Glycoproteomics Analysis in Blood and Other Bodily Fluids
An important target for blood-based diagnostic assays involves the detection and quantification of glycosylated proteins. Glycosylated proteins, especially N-linked glycoproteins, are ubiquitous among the proteins destined for extracellular environments (163), such as plasma or serum. A systematic and in-depth global profiling of the blood glycoproteome can provide fundamental knowledge for blood biomarker development, and is now possible with the development of glycoproteomics technologies. In the past few years, several large scale proteomics studies on profiling the glycoproteome of human plasma and serum have been reported (34, 35, 37, 38, 43, 61, 65, 164–166), adding significant numbers of glycoproteins into the blood glycoproteome database. In one study (38), immunoaffinity subtraction and hydrazide chemistry were applied to enrich N-glycoproteins from human plasma. The captured plasma glycoproteins were subjected to two-dimensional liquid chromatography separation followed by tandem mass spectrometric analysis. A total of 2053 different N-glycopeptides were identified, covering 303 nonredundant glycoproteins, including many glycoproteins with low abundance in blood (38). In a different study, hydrazide chemistry-based solid phase extraction method was applied to enhance the detection of tissue-derived proteins in human plasma (167). Other studies have applied lectin affinity-based approaches to characterize the serum and plasma glycoproteome (34, 43, 166). These studies provide detailed identification regarding the individual N-glycosylation sites using high-resolution mass spectrometry. The efforts made in global profiling of glycoproteins in plasma and serum have not only greatly enhanced our understanding of the blood glycoproteome, but also have facilitated the development of new technologies that can be used for glycoprotein biomarker discovery. A variety of experimental designs and strategies for blood glycoprotein profiling have been applied for clinical disease studies, including prostate cancer (168), hepatocellular carcinoma (164, 168–170), lung adenocarcinoma (61, 171), breast cancer (58, 165, 172), atopic dermatitis (169), ovarian cancer (173, 174), congenital disorders of glycosylation (175), and pancreatic cancer (156, 176). Most of these studies focused on the early stages of glycoprotein biomarker discovery and many of them exploited multilectin affinity techniques to isolate glycoproteins from serum or plasma.
Glycoproteomics techniques have also been applied to study the glycoproteome of other bodily fluids. The complementary application of hydrazide chemistry-based solid phase extraction and lectin affinity method have led to the identification of 216 glycoproteins in human cerebrospinal fluid (CSF), including many low abundant ones (39). A hydrazide chemistry based study on human saliva has characterized 84 N-glycosylated peptides in 45 glycoproteins (177). The study on tear fluid identified 43 N-linked glycoproteins, including 19 proteins that have not been discovered in tear fluid previously (178). Other glycoproteomics studies on bodily fluids include N-glycoprotein profiling of lung adenocarcinoma pleural effusions (179), urine glycoprotein profiling (180), and urine glycoprotein signature identification for bladder cancer (181). In the urine glycoprotein profiling study, 150 annotated glycoproteins in addition to 43 predicted glycoproteins were identified (180). In our own study, 48 glycoproteins have so far been identified in pancreatic juice (unpublished data), adding complementary information to the pancreatic juice protein database (182–184).
Glycoproteomics Analysis of Tissue and Cell Lysates
Protein glycosylation has been increasingly recognized as one of the prominent alterations involved in tumorigenesis, inflammation, and other disease states. The study of glycoproteins in cell and tissue carries great promise for defining biomarkers for diagnotic and therapeutic targets. The glycoproteomics studies in liver tissue (185, 186) and cell lines (187) have provided a fundamental understanding of the liver glycoproteome and identified protein candidates that are associated with highly metastatic liver cancer cells. In one of the studies, hydrazide chemistry and multiple enzyme digestion provided a complementary identification of 939 N-glycosylation sites covering 523 nonredundant glycoproteins in human liver tissue (185). Studies on ovarian cancer have focused on discovering putative glycoprotein biomarkers for improving diagnosis (173, 174) and therapeutic treatment (188). Glycoproteomics studies have also been carried out to study hepatocelluar carcinoma. Magnetic nanoparticle immobilized Concanavalin A was used to selectively enrich N-glycoproteins in a hepatocelluar carcinoma cell line leading to the identification of 184 glycosylation sites corresponding to 101 glycoproteins (189). In a different study, complementary methods of hydrophilic affinity and hydrazide chemistry were applied to investigate the secreted glycoproteins from a hepatocelluar carcinoma cell line, in which 300 different glycosylation sites within 194 glycoproteins were identified (190). While many of these studies focused on N-glycoproteins, mucin-type O-linked glycoproteins are the predominant forms of O-linked glycosylation and are difficult to analyze. A metabolic labeling method was developed to facilitate their identification in complex cell lysates using proteomic strategies (191).
Cell surface and membrane proteins are particularly appealing for biomarker discovery, and many of them are glycosylated proteins. Both hydrazide chemistry- and lectin affinity-based approaches have been applied to specifically study cell surface and membrane N-glycoproteins that are associated with diseases, including colon carcinoma (192), breast cancer (158), and thyroid cancer (157). One study applied hydrazide chemistry to covalently label extracellular glycan moieties on live cells, providing highly specific and selective identification of cell surface N-glycoproteins (64). A complementary application of hydrazide chemistry and lectin affinity methods was demonstrated to profile cell membrane glycoproteins, significantly enhancing the glycoprotein identification (67).
Quantitative Glycoprotein Profiling
One of the major goals of clinical proteomics is to effectively identify dysregulated proteins that are specifically associated with a biological state, such as a disease. In the past decade, different quantitative proteomics techniques have been introduced and applied to study a wide variety of disease settings. These techniques are based on different mechanisms to facilitate mass spectrometric-based quantitative analysis, including stable isotopic or isobaric labeling using chemical reactions (e.g. ICAT and iTRAQ) (193–195), metabolic incorporation (e.g. SILAC) (196) and enzymatic reactions (e.g. 18O labeling) (197, 198); as well as less quantitatively accurate label-free approaches (199, 200). The overview and comparison of these quantitative techniques can be found in several reports in the literature and are not discussed in this review. Most of these isotopic labeling techniques can be adapted and utilized for glycoproteomics analysis to quantitatively compare the glycoproteome of a diseased sample to a control, thus revealing the glycosylation occupancy of individual glycosylation sites that may be involved in a disease. In addition to the well-established labeling methods cited above, several more experimental labeling strategies have been described in the field of glycoproteomics. One study demonstrated the feasibility of using stable isotope labeled succinic anhydride for quantitative analysis of glycoproteins isolated from serum via hydrazide chemistry (37). In a different report, the heavy and light version of N-acetoxy-succinimide combining with lectin affinity selection was used to quantitatively profile serum glycopeptides in canine lymphoma and transitional cell carcinoma (201). Stable isotope labeled 2-nitrobenzenesulfenyl was also used for chemical labeling in a quantitative glycoprotein profiling study on the sera from patients with lung adenocarcinoma (202). O-Linked N-acetylglucosamine (O-GlcNAc) is an intracellular, reversible form of glycosylation that shares many features with phosphorylation (203). Studies have suggested that O-GlcNAc may play an important role in many biological processes (204). A quantitative study on O-GlcNAc glycosylation has been reported, in which a method termed quantitative isotopic and chemoenzymatic tagging (QUIC-Tag) was described using a biotin-avidin affinity strategy for O-GlcNAc glycopeptide enrichment and stable isotope-labeled formaldehyde for mass spectrometric quantification (205). Recently, the isobaric tag for relative and absolute quantitation (iTRAQ) technique, combined with different glycoprotein enrichment approaches, has been utilized in several quantitative glycoproteomics studies. In the study of hepatocellular carcinoma, N-linked glycoproteins were enriched from hepatocellular carcinoma patients and controls using multilectin column and then quantitatively compared using iTRAQ to reveal the differential proteins associated with hepatocellular carcinoma (206). In a different study, the approach of using narrow selectivity lectin affinity chromatography followed by iTRAQ labeling was demonstrated to selectively identify differential glycoproteins in plasma samples from breast cancer patients (165). Another study utilized hydrazide chemistry-based solid phase extraction and iTRAQ to investigate the tear fluid of patients with climatic droplet keratopathy in comparison of normal controls, identifying multiple N-glycosylation sites with differential occupancy associated with climatic droplet keratopathy (178).
In addition to using chemical reactions to incorporate stable isotope tag for quantitative mass spectrometric analysis, 18O can be introduced into N-glycopeptides during enzymatic reactions, such as tryptic digestion (incorporation of two 18O into the peptide carboxyl-terminal) and PNGase F mediated hydrolysis (incorporation of one 18O into the asparagine of N-glycosylation sites (33)). Attempts have been made to apply this approach to identify differentially expressed N-glycosylation associated with ovarian cancer in serum (207). In a different approach, the SILAC technique allows incorporation of stable isotope-labeled amino acids into proteins during cell culturing process (196), and was applied to investigate the difference in cell surface N-glycoproteins among different cell types (64). A label-free approach has also been used for glycoproteomics profiling, including a method developed to profile intact glycopeptides in a complex sample (208) and a study that compares the plasma glycoproteome between psoriasis patients and healthy controls (209).
Targeted Glycoproteomics Analysis
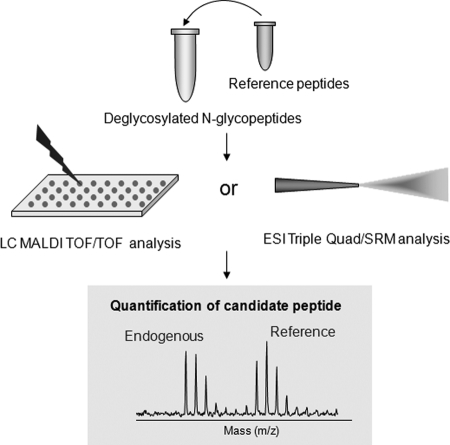
Mass spectrometry based targeted proteomics has recently emerged as a multiplexed quantitative technique that affords highly specific and candidate-based detection of targeted peptides and proteins in a complex biological sample (18, 210–214). The technique is based on the concept of stable isotope dilution utilizing stable isotope-labeled synthetic reference peptides, which precisely mimic their endogenous counterparts, to achieve targeted quantification (214). Such techniques can be applied to target specific glycoproteins or glycopeptides, to precisely quantify the status of candidate glycosylation sites and assess the glycosylation occupancy at the molecular level. However, it is technically impractical to use synthetic peptides to precisely mimic a large number of natural glycopeptides with intact a glycan moiety as internal standards because of the structure complexity and variation of the sugar chain. To overcome these technical obstacles, an alternative approach was proposed for targeted analysis of N-glycosylation occupancy, in which stable isotope-labeled peptides were synthesized to mimic the deglycosylated form of candidate glycopeptides as internal references (161). It is known that the deglycosylation step using PNGase F results in a conversion of asparagine to aspartic acid in the peptide sequence, introducing a mass difference of 0.9840 Da. This phenomenon was utilized to design a synthetic peptide to mimic the endogenous N-linked glycopeptide in its deglycosylation form with exact amino acid sequence of its endogenous counterpart and with 13C and 15N labeling on one of its amino acids (161). Therefore, each matched pair of reference and endogenous candidate glycopeptides should share the same chromatographic and mass spectrometric characteristics, and can only be distinguished by their mass difference and isotopic pattern because of isotopic labeling. This design conceptually ensures that the synthetic internal standard of a candidate glycopeptide will be detected simultaneously with its endogenous form under the same analytical conditions, thus, minimizing the systematic variation and providing reliable quantification (214). The strategy for targeted glycoproteomics analysis is schematically illustrated in Fig. 2.
Fig. 2.
Targeted analysis of N-glycopeptides.
The targeted glycoproteomics technique was first demonstrated to analyze N-glycopeptides that were extracted from human serum using an integrated pipeline combining a hydrazide chemistry-based solid phase extraction method and a data-driven liquid chromatography MALDI TOF/TOF mass spectrometric analysis to quantify 21 N-glycopeptides in human serum (161). A similar mass spectrometric platform was then applied in a different study to assess a subset of glycoprotein biomarker candidates in the sera from prostate cancer patients (215). The targeted glycoproteomics analysis has also been demonstrated using a triple Q/linear ion trap instrument with the selected reaction monitoring (also referred to as multiple reaction monitoring) technique for highly sensitive targeted detection of N-glycoproteins in plasma (159). The technique was applied to detect tissue inhibitor of metalloproteinase 1 (TIMP1), an aberrant glycoprotein associated with colorectal cancer, in the sera of colorectal cancer patients (216) using a tandem enrichment strategy, combing lectin glycoprotein enrichment followed by the method of stable isotope standards and capture by antipeptide antibodies (SISCAPA), to enhance the detection of tissue inhibitor of metalloproteinase 1 (216). These studies demonstrate an integrated pipeline for candidate-based glycoproteomics analysis with precise mapping of targeted N-linked motifs and absolute quantification of the glycoprotein targets in a complex biological sample. Such targeted glycoproteomics can reach a detection sensitivity at the nanogram per milliliter level for serum and plasma detection (159, 214–216).
Concluding Remarks
The major challenge for a comprehensive glycoproteomics analysis arises not only from the enormous complexity and nonlinear dynamic range in protein constituent in a clinical sample, but also the profound biological intricacy within the molecule of a glycoprotein, involving the flexibility in glycan structures and the complex linkage with the corresponding protein. In the past decade, significant efforts have been made to structurally or quantitatively characterize the glycoproteome of a variety of biological samples, and to investigate the significant glycoproteins in a wide assortment of diseases. Shotgun proteomics-based techniques are still the most effective and versatile approach in glycoproteomics analysis, allowing high throughput and detailed analysis on individual glycosylation sites. Although glycoproteomics is quickly emerging as an important technique for clinical proteomics study and biomarker discovery, a comprehensive, quantitative glycoproteomics analysis in a complex biological sample still remains challenging. It is anticipated that with the continued evolution in mass spectrometry, separation technology, and bioinformatics many of the technical limitations associated with current glycoproteomics may be transient. There is no doubt that glycoproteomics is playing an increasingly important role in biomarker discovery and clinical study.
Footnotes
* This work was supported in part with federal funds from the National Institutes of Health (K25CA137222, R01CA107209, K07CA116296, R01DK081368) and a grant from Canary Foundation (TAB). RA acknowledges funding from the Swiss National Science Foundation (3100A0-107679).
1 The abbreviations used are:
- CID
- collision-induced dissociation
- Q
- quadrupole.
REFERENCES
- 1. Cooper C. A., Harrison M. J., Wilkins M. R., Packer N. H. (2001) GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 29, 332–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rudd P. M., Dwek R. A. (1997) Glycosylation: heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 32, 1–100 [DOI] [PubMed] [Google Scholar]
- 3. Rudd P. M., Guile G. R., Küster B., Harvey D. J., Opdenakker G., Dwek R. A. (1997) Oligosaccharide sequencing technology. Nature 388, 205–207 [DOI] [PubMed] [Google Scholar]
- 4. Brooks S. A., Carter T. M., Royle L., Harvey D. J., Fry S. A., Kinch C., Dwek R. A., Rudd P. M. (2008) Altered glycosylation of proteins in cancer: what is the potential for new anti-tumour strategies. Anticancer Agents Med. Chem. 8, 2–21 [DOI] [PubMed] [Google Scholar]
- 5. Kobata A. (1989) Altered glycosylation of surface glycoproteins in tumor cells and its clinical application. Pigment Cell Res. 2, 304–308 [DOI] [PubMed] [Google Scholar]
- 6. Kobata A., Amano J. (2005) Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol. Cell Biol. 83, 429–439 [DOI] [PubMed] [Google Scholar]
- 7. Kam R. K. T., Poon T. C. W. (2008) The potentials of glycomics in biomarker discovery. Clin. Proteom. 4, 67–79 [Google Scholar]
- 8. Shriver Z., Raguram S., Sasisekharan R. (2004) Glycomics: a pathway to a class of new and improved therapeutics. Nat. Rev. Drug Discov. 3, 863–873 [DOI] [PubMed] [Google Scholar]
- 9. Sierpina V. S., Murray R. K. (2006) Glyconutrients: the state of the science and the impact of glycomics. Explore (NY) 2, 488–494 [DOI] [PubMed] [Google Scholar]
- 10. Taniguchi N. (2008) Toward cancer biomarker discovery using the glycomics approach. Proteomics 8, 3205–3208 [DOI] [PubMed] [Google Scholar]
- 11. Dennis J. W., Granovsky M., Warren C. E. (1999) Glycoprotein glycosylation and cancer progression. Biochim. Biophys. Acta 1473, 21–34 [DOI] [PubMed] [Google Scholar]
- 12. Ono M., Hakomori S. (2004) Glycosylation defining cancer cell motility and invasiveness. Glycoconj. J. 20, 71–78 [DOI] [PubMed] [Google Scholar]
- 13. Vlad A. M., Finn O. J. (2004) Glycoprotein tumor antigens for immunotherapy of breast cancer. Breast Dis. 20, 73–79 [DOI] [PubMed] [Google Scholar]
- 14. Ludwig J. A., Weinstein J. N. (2005) Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 5, 845–856 [DOI] [PubMed] [Google Scholar]
- 15. Polanski M., Anderson N. L. (2007) A list of candidate cancer biomarkers for targeted proteomics. Biomark. Insights. 1, 1–48 [PMC free article] [PubMed] [Google Scholar]
- 16. Hartwell L., Mankoff D., Paulovich A., Ramsey S., Swisher E. (2006) Cancer biomarkers: a systems approach. Nat. Biotechnol. 24, 905–908 [DOI] [PubMed] [Google Scholar]
- 17. Pepe M. S., Etzioni R., Feng Z., Potter J. D., Thompson M. L., Thornquist M., Winget M., Yasui Y. (2001) Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 93, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 18. Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 19. Bause E. (1983) Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem. J. 209, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei X., Li L. (2009) Comparative glycoproteomics: approaches and applications. Brief. Funct. Genomic. Proteomic. 8, 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Floyd N., Vijayakrishnan B., Koeppe J. R., Davis B. G. (2009) Thiyl glycosylation of olefinic proteins: S-linked glycoconjugate synthesis. Angew. Chem. Int. Ed Engl. 48, 7798–7802 [DOI] [PubMed] [Google Scholar]
- 22. Lote C. J., Weiss J. B. (1971) Identification in urine of a low-molecular-weight highly polar glycopeptide containing cysteinyl-galactose. Biochem. J. 123, 25P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 24. Aoki-Kinoshita K. F. (2008) An introduction to bioinformatics for glycomics research. PLoS. Comput. Biol. 4, e1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von der Lieth C. W., Lütteke T., Frank M. (2006) The role of informatics in glycobiology research with special emphasis on automatic interpretation of MS spectra. Biochim. Biophys. Acta 1760, 568–577 [DOI] [PubMed] [Google Scholar]
- 26. North S. J., Hitchen P. G., Haslam S. M., Dell A. (2009) Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr. Opin. Struct. Biol. 19, 498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tajiri M., Yoshida S., Wada Y. (2005) Differential analysis of site-specific glycans on plasma and cellular fibronectins: application of a hydrophilic affinity method for glycopeptide enrichment. Glycobiology 15, 1332–1340 [DOI] [PubMed] [Google Scholar]
- 29. Uematsu R., Furukawa J., Nakagawa H., Shinohara Y., Deguchi K., Monde K., Nishimura S. (2005) High throughput quantitative glycomics and glycoform-focused proteomics of murine dermis and epidermis. Mol. Cell Proteomics 4, 1977–1989 [DOI] [PubMed] [Google Scholar]
- 30. Wada Y., Tajiri M., Yoshida S. (2004) Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal. Chem. 76, 6560–6565 [DOI] [PubMed] [Google Scholar]
- 31. Wang Y., Wu S. L., Hancock W. S. (2006) Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap–Fourier transform mass spectrometry. Glycobiology 16, 514–523 [DOI] [PubMed] [Google Scholar]
- 32. Geng M., Zhang X., Bina M., Regnier F. (2001) Proteomics of glycoproteins based on affinity selection of glycopeptides from tryptic digests. J. Chromatogr. B Biomed. Sci. Appl. 752, 293–306 [DOI] [PubMed] [Google Scholar]
- 33. Kaji H., Saito H., Yamauchi Y., Shinkawa T., Taoka M., Hirabayashi J., Kasai K., Takahashi N., Isobe T. (2003) Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 21, 667–672 [DOI] [PubMed] [Google Scholar]
- 34. Yang Z., Hancock W. S. (2004) Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J. Chromatogr. A 1053, 79–88 [PubMed] [Google Scholar]
- 35. Durham M., Regnier F. E. (2006) Targeted glycoproteomics: serial lectin affinity chromatography in the selection of O-glycosylation sites on proteins from the human blood proteome. J. Chromatogr. A 1132, 165–173 [DOI] [PubMed] [Google Scholar]
- 36. Qiu R., Regnier F. E. (2005) Use of multidimensional lectin affinity chromatography in differential glycoproteomics. Anal. Chem. 77, 2802–2809 [DOI] [PubMed] [Google Scholar]
- 37. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 38. Liu T., Qian W. J., Gritsenko M. A., Camp D. G., Monroe M. E., Moore R. J., Smith R. D. (2005) Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 4, 2070–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pan S., Wang Y., Quinn J. F., Peskind E. R., Waichunas D., Wimberger J. T., Jin J., Li J. G., Zhu D., Pan C., Zhang J. (2006) Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J. Proteome Res. 5, 2769–2779 [DOI] [PubMed] [Google Scholar]
- 40. Zhang H., Aebersold R. (2006) Isolation of glycoproteins and identification of their N-linked glycosylation sites. Methods Mol. Biol. 328, 177–185 [DOI] [PubMed] [Google Scholar]
- 41. Sun B., Ranish J. A., Utleg A. G., White J. T., Yan X., Lin B., Hood L. (2007) Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol. Cell Proteomics 6, 141–149 [DOI] [PubMed] [Google Scholar]
- 42. Zhou Y., Aebersold R., Zhang H. (2007) Isolation of N-linked glycopeptides from plasma. Anal. Chem. 79, 5826–5837 [DOI] [PubMed] [Google Scholar]
- 43. Yang Z., Hancock W. S., Chew T. R., Bonilla L. (2005) A study of glycoproteins in human serum and plasma reference standards (HUPO) using multilectin affinity chromatography coupled with RPLC-MS/MS. Proteomics. 5, 3353–3366 [DOI] [PubMed] [Google Scholar]
- 44. Sparbier K., Koch S., Kessler I., Wenzel T., Kostrzewa M. (2005) Selective isolation of glycoproteins and glycopeptides for MALDI-TOF MS detection supported by magnetic particles. J. Biomol. Tech. 16, 407–413 [PMC free article] [PubMed] [Google Scholar]
- 45. Sparbier K., Wenzel T., Kostrzewa M. (2006) Exploring the binding profiles of ConA, boronic acid and WGA by MALDI-TOF/TOF MS and magnetic particles. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 840, 29–36 [DOI] [PubMed] [Google Scholar]
- 46. Alvarez-Manilla G., Atwood J., 3rd, Guo Y., Warren N.L., Orlando R., Pierce M. (2006) Tools for glycoproteomic analysis: size exclusion chromatography facilitates identification of tryptic glycopeptides with N-linked glycosylation sites. J. Proteome Res. 5, 701–708 [DOI] [PubMed] [Google Scholar]
- 47. Hägglund P., Bunkenborg J., Elortza F., Jensen O. N., Roepstorff P. (2004) A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 3, 556–566 [DOI] [PubMed] [Google Scholar]
- 48. Larsen M. R., Højrup P., Roepstorff P. (2005) Characterization of gel-separated glycoproteins using two-step proteolytic digestion combined with sequential microcolumns and mass spectrometry. Mol. Cell Proteomics 4, 107–119 [DOI] [PubMed] [Google Scholar]
- 49. Endo T. (1996) Fractionation of glycoprotein-derived oligosaccharides by affinity chromatography using immobilized lectin columns. J. Chromatogr. A 720, 251–261 [DOI] [PubMed] [Google Scholar]
- 50. Saleemuddin M., Husain Q. (1991) Concanavalin A: a useful ligand for glycoenzyme immobilization–a review. Enzyme Microb. Technol. 13, 290–295 [DOI] [PubMed] [Google Scholar]
- 51. Becker J. W., Reeke G. N., Jr., Wang J. L., Cunningham B. A., Edelman G. M. (1975) The covalent and three-dimensional structure of concanavalin A. Structure of the monomer and its interactions with metals and saccharides. J. Biol. Chem. 250, 1513–1524 [PubMed] [Google Scholar]
- 52. Bakry N., Kamata Y., Simpson L. L. (1991) Lectins from Triticum vulgaris and Limax flavus are universal antagonists of botulinum neurotoxin and tetanus toxin. J. Pharmacol. Exp. Ther. 258, 830–836 [PubMed] [Google Scholar]
- 53. Saulsbury F. T. (1997) Alterations in the O-linked glycosylation of IgA1 in children with Henoch-Schonlein purpura. J. Rheumatol. 24, 2246–2249 [PubMed] [Google Scholar]
- 54. Cummings R. D., Kornfeld S. (1982) Fractionation of asparagine-linked oligosaccharides by serial lectin-Agarose affinity chromatography. A rapid, sensitive, and specific technique. J. Biol. Chem. 257, 11235–11240 [PubMed] [Google Scholar]
- 55. Qiu R., Regnier F. E. (2005) Comparative glycoproteomics of N-linked complex-type glycoforms containing sialic acid in human serum. Anal. Chem. 77, 7225–7231 [DOI] [PubMed] [Google Scholar]
- 56. Wuhrer M., Catalina M. I., Deelder A. M., Hokke C. H. (2007) Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 849, 115–128 [DOI] [PubMed] [Google Scholar]
- 57. Wang Y., Wu S. L., Hancock W. S. (2006) Monitoring of glycoprotein products in cell culture lysates using lectin affinity chromatography and capillary HPLC coupled to electrospray linear ion trap-Fourier transform mass spectrometry (LTQ/FTMS). Biotechnol. Prog. 22, 873–880 [DOI] [PubMed] [Google Scholar]
- 58. Yang Z., Harris L. E., Palmer-Toy D. E., Hancock W. S. (2006) Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin. Chem. 52, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 59. Kullolli M., Hancock W. S., Hincapie M. (2008) Preparation of a high-performance multi-lectin affinity chromatography (HP-M-LAC) adsorbent for the analysis of human plasma glycoproteins. J. Sep. Sci. 31, 2733–2739 [DOI] [PubMed] [Google Scholar]
- 60. Bunkenborg J., Pilch B. J., Podtelejnikov A. V., Wiśniewski J. R. (2004) Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics 4, 454–465 [DOI] [PubMed] [Google Scholar]
- 61. Heo S. H., Lee S. J., Ryoo H. M., Park J. Y., Cho J. Y. (2007) Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics 7, 4292–4302 [DOI] [PubMed] [Google Scholar]
- 62. Yang Z., Hancock W. S. (2005) Monitoring glycosylation pattern changes of glycoproteins using multi-lectin affinity chromatography. J. Chromatogr. A 1070, 57–64 [DOI] [PubMed] [Google Scholar]
- 63. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 64. Wollscheid B., Bausch-Fluck D., Henderson C., O'Brien R., Bibel M., Schiess R., Aebersold R., Watts J. D. (2009) Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Calvano C. D., Zambonin C. G., Jensen O. N. (2008) Assessment of lectin and HILIC based enrichment protocols for characterization of serum glycoproteins by mass spectrometry. J. Proteomics 71, 304–317 [DOI] [PubMed] [Google Scholar]
- 66. Lee A., Kolarich D., Haynes P. A., Jensen P. H., Baker M. S., Packer N. H. (2009) Rat liver membrane glycoproteome: enrichment by phase partitioning and glycoprotein capture. J. Proteome Res. 8, 770–781 [DOI] [PubMed] [Google Scholar]
- 67. McDonald C. A., Yang J. Y., Marathe V., Yen T. Y., Macher B. A. (2009) Combining results from lectin affinity chromatography and glycocapture approaches substantially improves the coverage of the glycoproteome. Mol. Cell Proteomics 8, 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ahn B., Rhee S. G., Stadtman E. R. (1987) Use of fluorescein hydrazide and fluorescein thiosemicarbazide reagents for the fluorometric determination of protein carbonyl groups and for the detection of oxidized protein on polyacrylamide gels. Anal. Biochem. 161, 245–257 [DOI] [PubMed] [Google Scholar]
- 69. Meany D. L., Xie H., Thompson L. V., Arriaga E. A., Griffin T. J. (2007) Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics 7, 1150–1163 [DOI] [PubMed] [Google Scholar]
- 70. Mirzaei H., Regnier F. (2007) Identification of yeast oxidized proteins: chromatographic top-down approach for identification of carbonylated, fragmented and cross-linked proteins in yeast. J. Chromatogr. A 1141, 22–31 [DOI] [PubMed] [Google Scholar]
- 71. Rawn J. D., Lienhard G. E. (1974) The binding of boronic acids to chymotrypsin. Biochemistry 13, 3124–3130 [DOI] [PubMed] [Google Scholar]
- 72. Burlingame A. L. (1996) Characterization of protein glycosylation by mass spectrometry. Curr. Opin. Biotechnol. 7, 4–10 [DOI] [PubMed] [Google Scholar]
- 73. Harvey D. J. (2005) Proteomic analysis of glycosylation: structural determination of N- and O-linked glycans by mass spectrometry. Expert. Rev. Proteomics. 2, 87–101 [DOI] [PubMed] [Google Scholar]
- 74. Kelleher N. L. (2004) Top-down proteomics. Anal. Chem. 76, 197A–203A [PubMed] [Google Scholar]
- 75. Carr S. A., Huddleston M. J., Bean M. F. (1993) Selective identification and differentiation of N- and O-linked oligosaccharides in glycoproteins by liquid chromatography-mass spectrometry. Protein Sci. 2, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Huddleston M. J., Bean M. F., Carr S. A. (1993) Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal. Chem. 65, 877–884 [DOI] [PubMed] [Google Scholar]
- 77. Kieliszewski M. J., O'Neill M., Leykam J., Orlando R. (1995) Tandem mass spectrometry and structural elucidation of glycopeptides from a hydroxyproline-rich plant cell wall glycoprotein indicate that contiguous hydroxyproline residues are the major sites of hydroxyproline O-arabinosylation. J. Biol. Chem. 270, 2541–2549 [DOI] [PubMed] [Google Scholar]
- 78. Medzihradszky K. F., Gillece-Castro B. L., Settineri C. A., Townsend R. R., Masiarz F. R., Burlingame A. L. (1990) Structure determination of O-linked glycopeptides by tandem mass spectrometry. Biomed. Environ. Mass Spectrom. 19, 777–781 [DOI] [PubMed] [Google Scholar]
- 79. Settineri C. A., Medzihradszky K. F., Masiarz F. R., Burlingame A. L., Chu C., George-Nascimento C. (1990) Characterization of O-glycosylation sites in recombinant B-chain of platelet-derived growth factor expressed in yeast using liquid secondary ion mass spectrometry, tandem mass spectrometry and Edman sequence analysis. Biomed. Environ. Mass Spectrom. 19, 665–676 [DOI] [PubMed] [Google Scholar]
- 80. Odani H., Yamamoto K., Iwayama S., Iwase H., Takasaki A., Takahashi K., Fujita Y., Sugiyama S., Hiki Y. (2010) Evaluation of the specific structures of IgA1 hinge glycopeptide in 30 IgA nephropathy patients by mass spectrometry. J. Nephrol. 23, 70–76 [PubMed] [Google Scholar]
- 81. Wuhrer M., Koeleman C. A., Deelder A. M. (2009) Hexose rearrangements upon fragmentation of N-glycopeptides and reductively aminated N-glycans. Anal. Chem. 81, 4422–4432 [DOI] [PubMed] [Google Scholar]
- 82. Zhang L., Reilly J. P. (2009) Extracting both peptide sequence and glycan structural information by 157 nm photodissociation of N-linked glycopeptides. J. Proteome Res. 8, 734–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Temporini C., Perani E., Calleri E., Dolcini L., Lubda D., Caccialanza G., Massolini G. (2007) Pronase-immobilized enzyme reactor: an approach for automation in glycoprotein analysis by LC/LC-ESI/MSn. Anal. Chem. 79, 355–363 [DOI] [PubMed] [Google Scholar]
- 84. Sullivan B., Addona T. A., Carr S. A. (2004) Selective detection of glycopeptides on ion trap mass spectrometers. Anal. Chem. 76, 3112–3118 [DOI] [PubMed] [Google Scholar]
- 85. Catalina M. I., Koeleman C. A., Deelder A. M., Wuhrer M. (2007) Electron transfer dissociation of N-glycopeptides: loss of the entire N-glycosylated asparagine side chain. Rapid Commun. Mass Spectrom. 21, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 86. Demelbauer U. M., Zehl M., Plematl A., Allmaier G., Rizzi A. (2004) Determination of glycopeptide structures by multistage mass spectrometry with low-energy collision-induced dissociation: comparison of electrospray ionization quadrupole ion trap and matrix-assisted laser desorption/ionization quadrupole ion trap reflectron time-of-flight approaches. Rapid Commun. Mass Spectrom. 18, 1575–1582 [DOI] [PubMed] [Google Scholar]
- 87. Sandra K., Devreese B., Van Beeumen J., Stals I., Claeyssens M. (2004) The Q-Trap mass spectrometer, a novel tool in the study of protein glycosylation. J. Am. Soc. Mass Spectrom. 15, 413–423 [DOI] [PubMed] [Google Scholar]
- 88. Hashii N., Kawasaki N., Itoh S., Nakajima Y., Harazono A., Kawanishi T., Yamaguchi T. (2009) Identification of glycoproteins carrying a target glycan-motif by liquid chromatography/multiple-stage mass spectrometry: identification of Lewis x-conjugated glycoproteins in mouse kidney. J. Proteome Res. 8, 3415–3429 [DOI] [PubMed] [Google Scholar]
- 89. Peterman S. M., Mulholland J. J. (2006) A novel approach for identification and characterization of glycoproteins using a hybrid linear ion trap/FT-ICR mass spectrometer. J. Am. Soc. Mass Spectrom. 17, 168–179 [DOI] [PubMed] [Google Scholar]
- 90. Deguchi K., Ito H., Baba T., Hirabayashi A., Nakagawa H., Fumoto M., Hinou H., Nishimura S. (2007) Structural analysis of O-glycopeptides employing negative- and positive-ion multi-stage mass spectra obtained by collision-induced and electron-capture dissociations in linear ion trap time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 21, 691–698 [DOI] [PubMed] [Google Scholar]
- 91. Ito H., Takegawa Y., Deguchi K., Nagai S., Nakagawa H., Shinohara Y., Nishimura S. (2006) Direct structural assignment of neutral and sialylated N-glycans of glycopeptides using collision-induced dissociation MSn spectral matching. Rapid Commun. Mass Spectrom. 20, 3557–3565 [DOI] [PubMed] [Google Scholar]
- 92. Satomi Y., Shimonishi Y., Hase T., Takao T. (2004) Site-specific carbohydrate profiling of human transferrin by nano-flow liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 18, 2983–2988 [DOI] [PubMed] [Google Scholar]
- 93. Satomi Y., Shimonishi Y., Takao T. (2004) N-glycosylation at Asn(491) in the Asn-Xaa-Cys motif of human transferrin. FEBS Lett. 576, 51–56 [DOI] [PubMed] [Google Scholar]
- 94. Imre T., Schlosser G., Pocsfalvi G., Siciliano R., Molnár-Szöllosi E., Kremmer T., Malorni A., Vékey K. (2005) Glycosylation site analysis of human alpha-1-acid glycoprotein (AGP) by capillary liquid chromatography-electrospray mass spectrometry. J. Mass Spectrom. 40, 1472–1483 [DOI] [PubMed] [Google Scholar]
- 95. Harazono A., Kawasaki N., Itoh S., Hashii N., Ishii-Watabe A., Kawanishi T., Hayakawa T. (2006) Site-specific N-glycosylation analysis of human plasma ceruloplasmin using liquid chromatography with electrospray ionization tandem mass spectrometry. Anal. Biochem. 348, 259–268 [DOI] [PubMed] [Google Scholar]
- 96. Nemeth J. F., Hochgesang G. P., Jr., Marnett L. J., Caprioli R. M., Hochensang G.P., Jr. (2001) Characterization of the glycosylation sites in cyclooxygenase-2 using mass spectrometry. Biochemistry 40, 3109–3116 [DOI] [PubMed] [Google Scholar]
- 97. Henriksson H., Denman S. E., Campuzano I. D., Ademark P., Master E. R., Teeri T. T., Brumer H., 3rd (2003) N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochem. J. 375, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bykova N. V., Rampitsch C., Krokhin O., Standing K. G., Ens W. (2006) Determination and characterization of site-specific N-glycosylation using MALDI-Qq-TOF tandem mass spectrometry: case study with a plant protease. Anal. Chem. 78, 1093–1103 [DOI] [PubMed] [Google Scholar]
- 99. Krokhin O., Ens W., Standing K. G., Wilkins J., Perreault H. (2004) Site-specific N-glycosylation analysis: matrix-assisted laser desorption/ionization quadrupole-quadrupole time-of-flight tandem mass spectral signatures for recognition and identification of glycopeptides. Rapid Commun. Mass Spectrom. 18, 2020–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Krokhin O. V., Ens W., Standing K. G. (2005) MALDI QqTOF MS combined with off-line HPLC for characterization of protein primary structure and post-translational modifications. J. Biomol. Tech. 16, 429–440 [PMC free article] [PubMed] [Google Scholar]
- 101. Kubota K., Sato Y., Suzuki Y., Goto-Inoue N., Toda T., Suzuki M., Hisanaga S., Suzuki A., Endo T. (2008) Analysis of glycopeptides using lectin affinity chromatography with MALDI-TOF mass spectrometry. Anal. Chem. 80, 3693–3698 [DOI] [PubMed] [Google Scholar]
- 102. Suzuki Y., Suzuki M., Nakahara Y., Ito Y., Ito E., Goto N., Miseki K., Iida J., Suzuki A. (2006) Structural characterization of glycopeptides by N-terminal protein ladder sequencing. Anal. Chem. 78, 2239–2243 [DOI] [PubMed] [Google Scholar]
- 103. Wuhrer M., Hokke C. H., Deelder A. M. (2004) Glycopeptide analysis by matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry reveals novel features of horseradish peroxidase glycosylation. Rapid Commun. Mass Spectrom. 18, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 104. Irungu J., Go E. P., Zhang Y., Dalpathado D. S., Liao H. X., Haynes B. F., Desaire H. (2008) Comparison of HPLC/ESI-FTICR MS versus MALDI-TOF/TOF MS for glycopeptide analysis of a highly glycosylated HIV envelope glycoprotein. J. Am. Soc. Mass Spectrom. 19, 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sparbier K., Asperger A., Resemann A., Kessler I., Koch S., Wenzel T., Stein G., Vorwerg L., Suckau D., Kostrzewa M. (2007) Analysis of glycoproteins in human serum by means of glycospecific magnetic bead separation and LC-MALDI-TOF/TOF analysis with automated glycopeptide detection. J. Biomol. Tech. 18, 252–258 [PMC free article] [PubMed] [Google Scholar]
- 106. Domon B., Costello C. E. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 5, 397–409 [Google Scholar]
- 107. Kurogochi M., Nishimura S. (2004) Structural characterization of N-glycopeptides by matrix-dependent selective fragmentation of MALDI-TOF/TOF tandem mass spectrometry. Anal. Chem. 76, 6097–6101 [DOI] [PubMed] [Google Scholar]
- 108. Kurogochi M., Matsushita T., Nishimura S. (2004) Post-translational modifications on proteins: facile and efficient procedure for the identification of O-glycosylation sites by MALDI-LIFT-TOF/TOF mass spectrometry. Angew. Chem. Int. Ed Engl. 43, 4071–4075 [DOI] [PubMed] [Google Scholar]
- 109. Nishimura S., Niikura K., Kurogochi M., Matsushita T., Fumoto M., Hinou H., Kamitani R., Nakagawa H., Deguchi K., Miura N., Monde K., Kondo H. (2004) High-throughput protein glycomics: combined use of chemoselective glycoblotting and MALDI-TOF/TOF mass spectrometry. Angew. Chem. Int. Ed Engl. 44, 91–96 [DOI] [PubMed] [Google Scholar]
- 110. Takemori N., Komori N., Matsumoto H. (2006) Highly sensitive multistage mass spectrometry enables small-scale analysis of protein glycosylation from two-dimensional polyacrylamide gels. Electrophoresis 27, 1394–1406 [DOI] [PubMed] [Google Scholar]
- 111. Wuhrer M., Balog C. I., Koeleman C. A., Deelder A. M., Hokke C. H. (2005) New features of site-specific horseradish peroxidase (HRP) glycosylation uncovered by nano-LC-MS with repeated ion-isolation/fragmentation cycles. Biochim. Biophys. Acta 1723, 229–239 [DOI] [PubMed] [Google Scholar]
- 112. Adamson J. T., Håkansson K. (2006) Infrared multiphoton dissociation and electron capture dissociation of high-mannose type glycopeptides. J. Proteome Res. 5, 493–501 [DOI] [PubMed] [Google Scholar]
- 113. Håkansson K., Cooper H. J., Emmett M. R., Costello C. E., Marshall A. G., Nilsson C. L. (2001) Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal. Chem. 73, 4530–4536 [DOI] [PubMed] [Google Scholar]
- 114. Håkansson K., Chalmers M. J., Quinn J. P., McFarland M. A., Hendrickson C. L., Marshall A. G. (2003) Combined electron capture and infrared multiphoton dissociation for multistage MS/MS in a Fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 75, 3256–3262 [DOI] [PubMed] [Google Scholar]
- 115. Seipert R. R., Dodds E. D., Clowers B. H., Beecroft S. M., German J. B., Lebrilla C. B. (2008) Factors that influence fragmentation behavior of N-linked glycopeptide ions. Anal. Chem. 80, 3684–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Haselmann K. F., Budnik B. A., Olsen J. V., Nielsen M. L., Reis C. A., Clausen H., Johnsen A. H., Zubarev R. A. (2001) Advantages of external accumulation for electron capture dissociation in Fourier transform mass spectrometry. Anal. Chem. 73, 2998–3005 [DOI] [PubMed] [Google Scholar]
- 117. Kjeldsen F., Haselmann K. F., Budnik B. A., Sørensen E. S., Zubarev R. A. (2003) Complete characterization of posttranslational modification sites in the bovine milk protein PP3 by tandem mass spectrometry with electron capture dissociation as the last stage. Anal. Chem. 75, 2355–2361 [DOI] [PubMed] [Google Scholar]
- 118. Mirgorodskaya E., Roepstorff P., Zubarev R. A. (1999) Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal. Chem. 71, 4431–4436 [DOI] [PubMed] [Google Scholar]
- 119. Mormann M., Paulsen H., Peter-Katalinic J. (2005) Electron capture dissociation of O-glycosylated peptides: radical site-induced fragmentation of glycosidic bonds. Eur. J. Mass Spectrom. (Chichester, Eng) 11, 497–511 [DOI] [PubMed] [Google Scholar]
- 120. Renfrow M. B., Cooper H. J., Tomana M., Kulhavy R., Hiki Y., Toma K., Emmett M. R., Mestecky J., Marshall A. G., Novak J. (2005) Determination of aberrant O-glycosylation in the IgA1 hinge region by electron capture dissociation fourier transform-ion cyclotron resonance mass spectrometry. J. Biol. Chem. 280, 19136–19145 [DOI] [PubMed] [Google Scholar]
- 121. Snovida S. I., Bodnar E. D., Viner R., Saba J., Perreault H. (2010) A simple cellulose column procedure for selective enrichment of glycopeptides and characterization by nano LC coupled with electron-transfer and high-energy collisional-dissociation tandem mass spectrometry. Carbohydr. Res. 345, 792–801 [DOI] [PubMed] [Google Scholar]
- 122. Darula Z., Medzihradszky K. F. (2009) Affinity enrichment and characterization of mucin core-1 type glycopeptides from bovine serum. Mol. Cell Proteomics 8, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Alley W. R., Jr., Mechref Y., Novotny M. V. (2009) Characterization of glycopeptides by combining collision-induced dissociation and electron-transfer dissociation mass spectrometry data. Rapid Commun. Mass Spectrom. 23, 161–170 [DOI] [PubMed] [Google Scholar]
- 124. Dalpathado D. S., Desaire H. (2008) Glycopeptide analysis by mass spectrometry. Analyst 133, 731–738 [DOI] [PubMed] [Google Scholar]
- 125. Jiang H., Desaire H., Butnev V. Y., Bousfield G. R. (2004) Glycoprotein profiling by electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 15, 750–758 [DOI] [PubMed] [Google Scholar]
- 126. Liu T., Li J. D., Zeng R., Shao X. X., Wang K. Y., Xia Q. C. (2001) Capillary electrophoresis-electrospray mass spectrometry for the characterization of high-mannose-type N-glycosylation and differential oxidation in glycoproteins by charge reversal and protease/glycosidase digestion. Anal. Chem. 73, 5875–5885 [DOI] [PubMed] [Google Scholar]
- 127. Wang F., Nakouzi A., Angeletti R. H., Casadevall A. (2003) Site-specific characterization of the N-linked oligosaccharides of a murine immunoglobulin M by high-performance liquid chromatography/electrospray mass spectrometry. Anal. Biochem. 314, 266–280 [DOI] [PubMed] [Google Scholar]
- 128. Dalpathado D. S., Irungu J., Go E. P., Butnev V. Y., Norton K., Bousfield G. R., Desaire H. (2006) Comparative glycomics of the glycoprotein follicle stimulating hormone: glycopeptide analysis of isolates from two mammalian species. Biochemistry 45, 8665–8673 [DOI] [PubMed] [Google Scholar]
- 129. Irungu J., Dalpathado D. S., Go E. P., Jiang H., Ha H. V., Bousfield G. R., Desaire H. (2006) Method for characterizing sulfated glycoproteins in a glycosylation site-specific fashion, using ion pairing and tandem mass spectrometry. Anal. Chem. 78, 1181–1190 [DOI] [PubMed] [Google Scholar]
- 130. Kjeldsen F., Haselmann K. F., Budnik B. A., Sørensen E. S., Zubarev R. A. (2003) Complete characterization of posttranslational modification sites in the bovine milk protein PP3 by tandem mass spectrometry with electron capture dissociation as the last stage. Anal. Chem. 75, 2355–2361 [DOI] [PubMed] [Google Scholar]
- 131. Chalabi S., Panico M., Sutton-Smith M., Haslam S. M., Patankar M. S., Lattanzio F. A., Morris H. R., Clark G. F., Dell A. (2006) Differential O-glycosylation of a conserved domain expressed in murine and human ZP3. Biochemistry 45, 637–647 [DOI] [PubMed] [Google Scholar]
- 132. Chalkley R. J., Burlingame A. L. (2001) Identification of GlcNAcylation sites of peptides and alpha-crystallin using Q-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 12, 1106–1113 [DOI] [PubMed] [Google Scholar]
- 133. Hanisch F. G., Green B. N., Bateman R., Peter-Katalinic J. (1998) Localization of O-glycosylation sites of MUC1 tandem repeats by QTOF ESI mass spectrometry. J. Mass Spectrom. 33, 358–362 [DOI] [PubMed] [Google Scholar]
- 134. Macek B., Hofsteenge J., Peter-Katalinić J. (2001) Direct determination of glycosylation sites in O-fucosylated glycopeptides using nano-electrospray quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 15, 771–777 [DOI] [PubMed] [Google Scholar]
- 135. Müller S., Goletz S., Packer N., Gooley A., Lawson A. M., Hanisch F. G. (1997) Localization of O-glycosylation sites on glycopeptide fragments from lactation-associated MUC1. All putative sites within the tandem repeat are glycosylation targets in vivo. J. Biol. Chem. 272, 24780–24793 [DOI] [PubMed] [Google Scholar]
- 136. Müller S., Alving K., Peter-Katalinic J., Zachara N., Gooley A. A., Hanisch F. G. (1999) High density O-glycosylation on tandem repeat peptide from secretory MUC1 of T47D breast cancer cells. J. Biol. Chem. 274, 18165–18172 [DOI] [PubMed] [Google Scholar]
- 137. Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 138. Clowers B. H., Dodds E. D., Seipert R. R., Lebrilla C. B. (2007) Site determination of protein glycosylation based on digestion with immobilized nonspecific proteases and Fourier transform ion cyclotron resonance mass spectrometry. J. Proteome Res. 6, 4032–4040 [DOI] [PubMed] [Google Scholar]
- 139. Renfrow M. B., Mackay C. L., Chalmers M. J., Julian B. A., Mestecky J., Kilian M., Poulsen K., Emmett M. R., Marshall A. G., Novak J. (2007) Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Anal. Bioanal. Chem. 389, 1397–1407 [DOI] [PubMed] [Google Scholar]
- 140. Sihlbom C., van Dijk Härd I, Lidell M. E., Noll T., Hansson G. C., Bäckström M. (2009) Localization of O-glycans in MUC1 glycoproteins using electron-capture dissociation fragmentation mass spectrometry. Glycobiology 19, 375–381 [DOI] [PubMed] [Google Scholar]
- 141. Joenväärä S., Ritamo I., Peltoniemi H., Renkonen R. (2008) N-glycoproteomics - an automated workflow approach. Glycobiology 18, 339–349 [DOI] [PubMed] [Google Scholar]
- 142. Xiong L., Regnier F. E. (2002) Use of a lectin affinity selector in the search for unusual glycosylation in proteomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 782, 405–418 [DOI] [PubMed] [Google Scholar]
- 143. Atwood J. A., 3rd, Minning T., Ludolf F., Nuccio A., Weatherly D. B., Alvarez-Manilla G., Tarleton R., Orlando R. (2006) Glycoproteomics of Trypanosoma cruzi trypomastigotes using subcellular fractionation, lectin affinity, and stable isotope labeling. J. Proteome Res. 5, 3376–3384 [DOI] [PubMed] [Google Scholar]
- 144. Edge A. S. (2003) Deglycosylation of glycoproteins with trifluoromethanesulphonic acid: elucidation of molecular structure and function. Biochem. J. 376 (Pt 2), 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gerken T. A., Gupta R., Jentoft N. (1992) A novel approach for chemically deglycosylating O-linked glycoproteins. The deglycosylation of submaxillary and respiratory mucins. Biochemistry 31, 639–648 [DOI] [PubMed] [Google Scholar]
- 146. Greis K. D., Hayes B. K., Comer F. I., Kirk M., Barnes S., Lowary T. L., Hart G. W. (1996) Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry. Anal. Biochem. 234, 38–49 [DOI] [PubMed] [Google Scholar]
- 147. Hong J. C., Kim Y. S. (2000) Alkali-catalyzed beta-elimination of periodate-oxidized glycans: a novel method of chemical deglycosylation of mucin gene products in paraffin embedded sections. Glycoconj. J. 17, 691–703 [DOI] [PubMed] [Google Scholar]
- 148. Eng J., McCormack A. L., Yates J. R., 3rd (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 149. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 150. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 151. Hernandez P., Müller M., Appel R. D. (2006) Automated protein identification by tandem mass spectrometry: issues and strategies. Mass Spectrom. Rev. 25, 235–254 [DOI] [PubMed] [Google Scholar]
- 152. Nesvizhskii A. I., Aebersold R. (2004) Analysis, statistical validation and dissemination of large-scale proteomics datasets generated by tandem MS. Drug Discov. Today 9, 173–181 [DOI] [PubMed] [Google Scholar]
- 153. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 154. Nesvizhskii A. I. (2007) Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol. Biol. 367, 87–119 [DOI] [PubMed] [Google Scholar]
- 155. Zhang H., Yi E. C., Li X. J., Mallick P., Kelly-Spratt K. S., Masselon C. D., Camp D. G., Smith R. D., Kemp C. J., Aebersold R. (2005) High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol. Cell Proteomics 4, 144–155 [DOI] [PubMed] [Google Scholar]
- 156. Zhao J., Simeone D. M., Heidt D., Anderson M. A., Lubman D. M. (2006) Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: application to pancreatic cancer serum. J. Proteome Res. 5, 1792–1802 [DOI] [PubMed] [Google Scholar]
- 157. Arcinas A., Yen T. Y., Kebebew E., Macher B. A. (2009) Cell surface and secreted protein profiles of human thyroid cancer cell lines reveal distinct glycoprotein patterns. J. Proteome Res. 8, 3958–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Whelan S. A., Lu M., He J., Yan W., Saxton R. E., Faull K. F., Whitelegge J. P., Chang H. R. (2009) Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. J. Proteome Res. 8, 4151–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Stahl-Zeng J., Lange V., Ossola R., Eckhardt K., Krek W., Aebersold R., Domon B. (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
- 160. Schulz B. L., Aebi M. (2009) Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol. Cell Proteomics 8, 357–364 [DOI] [PubMed] [Google Scholar]
- 161. Pan S., Zhang H., Rush J., Eng J., Zhang N., Patterson D., Comb M. J., Aebersold R. H. (2005) High-throughput proteome-screening approach for biomarker detection. Mol. Cell Proteomics 4, 182–190 [DOI] [PubMed] [Google Scholar]
- 162. Fenn L. S., McLean J. A. (2009) Simultaneous glycoproteomics on the basis of structure using ion mobility-mass spectrometry. Mol. Biosyst. 5, 1298–1302 [DOI] [PubMed] [Google Scholar]
- 163. Roth J. (2002) Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem. Rev. 102, 285–303 [DOI] [PubMed] [Google Scholar]
- 164. Jia W., Lu Z., Fu Y., Wang H. P., Wang L. H., Chi H., Yuan Z. F., Zheng Z. B., Song L. N., Han H. H., Liang Y. M., Wang J. L., Cai Y., Zhang Y. K., Deng Y. L., Ying W. T., He S. M., Qian X. H. (2009) A strategy for precise and large scale identification of core fucosylated glycoproteins. Mol. Cell Proteomics 8, 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Jung K., Cho W., Regnier F. E. (2009) Glycoproteomics of plasma based on narrow selectivity lectin affinity chromatography. J. Proteome Res. 8, 643–650 [DOI] [PubMed] [Google Scholar]
- 166. Madera M., Mechref Y., Klouckova I., Novotny M. V. (2006) Semiautomated high-sensitivity profiling of human blood serum glycoproteins through lectin preconcentration and multidimensional chromatography/tandem mass spectrometry. J. Proteome Res. 5, 2348–2363 [DOI] [PubMed] [Google Scholar]
- 167. Zhang H., Liu A. Y., Loriaux P., Wollscheid B., Zhou Y., Watts J. D., Aebersold R. (2007) Mass spectrometric detection of tissue proteins in plasma. Mol. Cell Proteomics 6, 64–71 [DOI] [PubMed] [Google Scholar]
- 168. Drake R. R., Schwegler E. E., Malik G., Diaz J., Block T., Mehta A., Semmes O. J. (2006) Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol. Cell Proteomics 5, 1957–1967 [DOI] [PubMed] [Google Scholar]
- 169. Kim W. K., Hwang H. R., Kim do H., Lee P. Y., In Y. J., Ryu H. Y., Park S. G., Bae K. H., Lee S. C. (2008) Glycoproteomic analysis of plasma from patients with atopic dermatitis: CD5L and ApoE as potential biomarkers. Exp. Mol. Med. 40, 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Liu Y., He J., Li C., Benitez R., Fu S., Marrero J., Lubman D. M. (2010) Identification and confirmation of biomarkers using an integrated platform for quantitative analysis of glycoproteins and their glycosylations. J. Proteome Res. 9, 798–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Hongsachart P., Huang-Liu R., Sinchaikul S., Pan F. M., Phutrakul S., Chuang Y. M., Yu C. J., Chen S. T. (2009) Glycoproteomic analysis of WGA-bound glycoprotein biomarkers in sera from patients with lung adenocarcinoma. Electrophoresis 30, 1206–1220 [DOI] [PubMed] [Google Scholar]
- 172. Zeng Z., Hincapie M., Haab B. B., Hanash S., Pitteri S. J., Kluck S., Hogan J. M., Kennedy J., Hancock W. S. (2010) The development of an integrated platform to identify breast cancer glycoproteome changes in human serum. J. Chromatogr. A 1217, 3307–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Li B., An H. J., Kirmiz C., Lebrilla C. B., Lam K. S., Miyamoto S. (2008) Glycoproteomic analyses of ovarian cancer cell lines and sera from ovarian cancer patients show distinct glycosylation changes in individual proteins. J. Proteome Res. 7, 3776–3788 [DOI] [PubMed] [Google Scholar]
- 174. Abbott K. L., Lim J. M., Wells L., Benigno B. B., McDonald J. F., Pierce M. (2010) Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 10, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Sturiale L., Barone R., Palmigiano A., Ndosimao C. N., Briones P., Adamowicz M., Jaeken J., Garozzo D. (2008) Multiplexed glycoproteomic analysis of glycosylation disorders by sequential yolk immunoglobulins immunoseparation and MALDI-TOF MS. Proteomics 8, 3822–3832 [DOI] [PubMed] [Google Scholar]
- 176. Zhao J., Qiu W., Simeone D. M., Lubman D. M. (2007) N-linked. glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J. Proteome Res. 6, 1126–1138 [DOI] [PubMed] [Google Scholar]
- 177. Ramachandran P., Boontheung P., Xie Y., Sondej M., Wong D. T., Loo J. A. (2006) Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 5, 1493–1503 [DOI] [PubMed] [Google Scholar]
- 178. Lei Z., Beuerman R. W., Chew A. P., Koh S. K., Cafaro T. A., Urrets-Zavalia E. A., Urrets-Zavalia J. A., Li S. F., Serra H. M. (2009) Quantitative analysis of N-linked glycoproteins in tear fluid of climatic droplet keratopathy by glycopeptide capture and iTRAQ. J. Proteome Res. 8, 1992–2003 [DOI] [PubMed] [Google Scholar]
- 179. Soltermann A., Ossola R., Kilgus-Hawelski S., von Eckardstein A., Suter T., Aebersold R., Moch H. (2008) N-glycoprotein profiling of lung adenocarcinoma pleural effusions by shotgun proteomics. Cancer 114, 124–133 [DOI] [PubMed] [Google Scholar]
- 180. Wang L., Li F., Sun W., Wu S., Wang X., Zhang L., Zheng D., Wang J., Gao Y. (2006) Concanavalin A-captured glycoproteins in healthy human urine. Mol. Cell Proteomics 5, 560–562 [DOI] [PubMed] [Google Scholar]
- 181. Kreunin P., Zhao J., Rosser C., Urquidi V., Lubman D. M., Goodison S. (2007) Bladder cancer associated glycoprotein signatures revealed by urinary proteomic profiling. J. Proteome Res. 6, 2631–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Chen R., Pan S., Yi E. C., Donohoe S., Bronner M. P., Potter J. D., Goodlett D. R., Aebersold R., Brentnall T. A. (2006) Quantitative proteomic profiling of pancreatic cancer juice. Proteomics 6, 3871–3879 [DOI] [PubMed] [Google Scholar]
- 183. Chen R., Pan S., Cooke K., Moyes K. W., Bronner M. P., Goodlett D. R., Aebersold R., Brentnall T. A. (2007) Comparison of pancreas juice proteins from cancer versus pancreatitis using quantitative proteomic analysis. Pancreas 34, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Grønborg M., Bunkenborg J., Kristiansen T. Z., Jensen O. N., Yeo C. J., Hruban R. H., Maitra A., Goggins M. G., Pandey A. (2004) Comprehensive proteomic analysis of human pancreatic juice. J. Proteome Res. 3, 1042–1055 [DOI] [PubMed] [Google Scholar]
- 185. Chen R., Jiang X., Sun D., Han G., Wang F., Ye M., Wang L., Zou H. (2009) Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 8, 651–661 [DOI] [PubMed] [Google Scholar]
- 186. Sun Q., Liu Y., Lu W., Cheng G., Zhou H., Zhou X., Wei L., Dai Z., Guo K., Lu H. (2007) Con A affinity glycoproteomics of normal human liver tissue. Sci. China C. Life Sci. 50, 403–411 [DOI] [PubMed] [Google Scholar]
- 187. Xu Z., Zhou X., Lu H., Wu N., Zhao H., Zhang L., Zhang W., Liang Y. L., Wang L., Liu Y., Yang P., Zha X. (2007) Comparative glycoproteomics based on lectins affinity capture of N-linked glycoproteins from human Chang liver cells and MHCC97-H cells. Proteomics 7, 2358–2370 [DOI] [PubMed] [Google Scholar]
- 188. Di Michele M., Marcone S., Cicchillitti L., Della, Corte A., Ferlini C., Scambia G., Donati M. B., Rotilio D. (2010) Glycoproteomics of paclitaxel resistance in human epithelial ovarian cancer cell lines: towards the identification of putative biomarkers. J. Proteomics 73, 879–898 [DOI] [PubMed] [Google Scholar]
- 189. Tang J., Liu Y., Yin P., Yao G., Yan G., Deng C., Zhang X. (2010) Concanavalin A-immobilized magnetic nanoparticles for selective enrichment of glycoproteins and application to glycoproteomics in hepatocelluar carcinoma cell line. Proteomics 10, 2000–2014 [DOI] [PubMed] [Google Scholar]
- 190. Cao J., Shen C., Wang H., Shen H., Chen Y., Nie A., Yan G., Lu H., Liu Y., Yang P. (2009) Identification of N-glycosylation sites on secreted proteins of human hepatocellular carcinoma cells with a complementary proteomics approach. J. Proteome Res. 8, 662–672 [DOI] [PubMed] [Google Scholar]
- 191. Hang H. C., Yu C., Kato D. L., Bertozzi C. R. (2003) A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl Acad. Sci. 100, 14846–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Vercoutter-Edouart A. S., Slomianny M. C., Dekeyzer-Beseme O., Haeuw J. F., Michalski J. C. (2008) Glycoproteomics and glycomics investigation of membrane N-glycosylproteins from human colon carcinoma cells. Proteomics 8, 3236–3256 [DOI] [PubMed] [Google Scholar]
- 193. Chakraborty A., Regnier F. E. (2002) Global internal standard technology for comparative proteomics. J. Chromatogr. A 949, 173–184 [DOI] [PubMed] [Google Scholar]
- 194. Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 195. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 196. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 197. Mirgorodskaya O. A., Kozmin Y. P., Titov M. I., Körner R., Sönksen C. P., Roepstorff P. (2000) Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using (18)O-labeled internal standards. Rapid Commun. Mass Spectrom. 14, 1226–1232 [DOI] [PubMed] [Google Scholar]
- 198. Yao X., Freas A., Ramirez J., Demirev P. A., Fenselau C. (2001) Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal. Chem. 73, 2836–2842 [DOI] [PubMed] [Google Scholar]
- 199. Bondarenko P. V., Chelius D., Shaler T. A. (2002) Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Chem. 74, 4741–4749 [DOI] [PubMed] [Google Scholar]
- 200. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 201. Wilson C. R., Regnier F. E., Knapp D. W., Raskin R. E., Andrews D. A., Hooser S. B. (2008) Glycoproteomic profiling of serum peptides in canine lymphoma and transitional cell carcinoma. Vet. Comp Oncol. 6, 171–181 [DOI] [PubMed] [Google Scholar]
- 202. Ueda K., Katagiri T., Shimada T., Irie S., Sato T. A., Nakamura Y., Daigo Y. (2007) Comparative profiling of serum glycoproteome by sequential purification of glycoproteins and 2-nitrobenzensulfenyl (NBS) stable isotope labeling: a new approach for the novel biomarker discovery for cancer. J. Proteome Res. 6, 3475–3483 [DOI] [PubMed] [Google Scholar]
- 203. Zeidan Q., Hart G. W. (2010) The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 123 (Pt 1), 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Butkinaree C., Park K., Hart G. W. (2010) O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., Hsieh-Wilson L. C. (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 [DOI] [PubMed] [Google Scholar]
- 206. Lee H. J., Na K., Choi E. Y., Kim K. S., Kim H., Paik Y. K. (2010) Simple method for quantitative analysis of N-linked glycoproteins in hepatocellular carcinoma specimens. J. Proteome Res. 9, 308–318 [DOI] [PubMed] [Google Scholar]
- 207. Liu Z., Cao J., He Y., Qiao L., Xu C., Lu H., Yang P. (2010) Tandem 18O stable isotope labeling for quantification of N-glycoproteome. J. Proteome Res. 9, 227–236 [DOI] [PubMed] [Google Scholar]
- 208. Rebecchi K. R., Wenke J. L., Go E. P., Desaire H. (2009) Label-free quantitation: a new glycoproteomics approach. J. Am. Soc. Mass Spectrom. 20, 1048–1059 [DOI] [PubMed] [Google Scholar]
- 209. Plavina T., Wakshull E., Hancock W. S., Hincapie M. (2007) Combination of abundant protein depletion and multi-lectin affinity chromatography (M-LAC) for plasma protein biomarker discovery. J. Proteome Res. 6, 662–671 [DOI] [PubMed] [Google Scholar]
- 210. Aebersold R. (2003) Constellations in a cellular universe. Nature 422, 115–116 [DOI] [PubMed] [Google Scholar]
- 211. Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 212. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 213. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Pan S., Aebersold R., Chen R., Rush J., Goodlett D. R., McIntosh M. W., Zhang J., Brentnall T. A. (2009) Mass spectrometry based targeted protein quantification: methods and applications. J. Proteome Res. 8, 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Li Y., Sokoll L. J., Rush J., Meany D., Zou N., Chan D. W., Zhang H. (2009) Targeted detection of prostate cancer proteins in serum using heavy peptide standards and MALDI-TOF/TOF. Proteomics-Clin. Appl. 3, 597–608 [Google Scholar]
- 216. Ahn Y. H., Lee J. Y., Lee J. Y., Kim Y. S., Ko J. H., Yoo J. S. (2009) Quantitative analysis of an aberrant glycoform of TIMP1 from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J. Proteome Res. 8, 4216–4224 [DOI] [PubMed] [Google Scholar]