Abstract
Regular (phospho)lipid bilayers do not pose efficient barriers for the transport of hydrophobic molecules. The outer membrane (OM) surrounding Gram-negative bacteria is a non-typical, asymmetric bilayer with an outer layer of lipopolysaccharide (LPS). The sugar molecules of the LPS layer prevent spontaneous diffusion of hydrophobic molecules across the OM. Since regular OM channels such as porins do not allow passage of hydrophobic molecules, specialized OM transport proteins are required for their uptake. Such proteins, exemplified by channels of the FadL family, transport their substrates according to a lateral diffusion mechanism. Here, substrates diffuse from the lumen of the β-barrel laterally into the OM, via a stable opening in the wall of the barrel. In this way, the lipopolysaccharide barrier is bypassed and, by depositing the substrates into the OM, a driving force for uptake is provided. Lateral diffusion through protein channel walls also occurs in α-helical inner membrane proteins, and may represent a widespread mechanism for proteins that transport and interact with hydrophobic substrates.
Keywords: Outer membrane transport, lipopolysaccharide, FadL, hydrophobic molecules, lateral diffusion
1. Introduction
Biological membranes (biomembranes) are crucial for all forms of life. They enclose cells and their organelles and maintain the essential differences between the different compartments. All biomembranes are lipid bilayers, composed of two monolayers of lipid molecules that are amphiphilic, i.e. they possess both hydrophilic ("water-loving") and hydrophobic ("water-fearing") properties. Within the lipid bilayer, the hydrophobic moieties of the lipid molecules contact each other to form a hydrophobic core, whereas the hydrophilic head group moieties are exposed to water on both sides of the membrane. The hydrophobic core of the membrane serves as a very efficient barrier against the permeation of hydrophilic molecules, necessitating membrane proteins to transport polar solutes across the membrane and to mediate signaling between both sides of the membrane. Conversely, most biomembranes do not pose significant barriers for the passage of hydrophobic molecules such as long-chain fatty acids (LCFAs) and xenobiotics destined for biodegradation. Indeed, the work by Hamilton and colleagues has convincingly shown that LCFAs diffuse rapidly across regular lipid bilayers.[1,2] One reason for the fact that hydrophobic molecules readily diffuse through lipid bilayers is that phospholipids, the principal components of most lipid bilayers, have relatively small hydrophilic head groups. Moreover, the associations between the individual membrane lipids are generally not very strong, giving most biomembranes a fluid-like character.
Gram-negative bacteria such as Escherichia coli are surrounded by an envelope consisting of two membranes: an inner membrane (IM), which is a regular phospholipids bilayer, and an outer membrane (OM). Both membranes are separated by the aqueous periplasmic space (Scheme 1). The OM, which serves as the permeability barrier to the outside environment, is strikingly asymmetric, with an inner layer of phospholipids and an outer layer of lipopolysaccharide (LPS). Like all membrane lipids, LPS has an amphiphilic character. In this case, the hydrophobic moiety is lipid A, a glycolipid that has typically six or seven acyl chains (Scheme 2). Attached to lipid A are a large number of different sugars, forming the core oligosaccharides and the highly variable O-antigen oligosaccharides (the LPS of some bacteria, such as E. coli K12, lacks O-antigen). Many of the sugar moieties have carboxyl and phosphate groups that are linked by divalent metal ions.[3] The sugar layer on the outside of the cell, which can be over 30 Å thick, is the principal reason why the OM forms a very efficient barrier against the permeation of small hydrophobic molecules. The importance of LPS as a permeability barrier is clear from the fact that bacteria with defects in LPS biosynthesis are much more susceptible to various hydrophobic drugs.[3] Thus, the bacterial OM is an atypical membrane in the sense that it forms an efficient barrier for both hydrophilic and hydrophobic molecules. In the case of hydrophobic molecules, the principal barrier for OM passage consists of the thick polar sugar layer of the LPS.
Scheme 1.

Schematic representation of the Gram-negative bacterial cell envelope. The sugar portion of the LPS layer is colored red, and phospholipid head groups are colored green. OM, outer membrane; IM, inner membrane.
Scheme 2.

The structure of lipolopysaccharide (LPS). A) General structure of Lipid A. B) Schematic representation of the Lipid A, core and O-antigen regions of LPS. The O-antigen region consists of repeats that can be much longer than shown here.
2. OM transport proteins
In order to acquire the necessary ions and nutrients required for growth and function, the OM contains transport proteins, virtually all of which form β-barrels. These proteins can be divided into three classes, based on the way they transport their substrates.[3] The first class consists of the porins, such as E. coli OmpC and OmpF. These abundant proteins do not bind their substrates, but instead form water-filled channels in the OM that mediate the passive diffusion of small (Mw < 600 Da) hydrophilic molecules according to their concentration gradient. The second class of OM transport proteins consists of the substrate-specific channels (e.g. the E. coli sugar channel LamB). These proteins contain binding sites for their substrates, and are more efficient compared to porins when the external substrate concentrations are low. As is the case for porins, the substrate-specific channels also transport their substrates by diffusion. Members of the third major class of OM transport proteins, the TonB-dependent receptors, are quite distinct from both porins and substrate specific channels. TonB-dependent receptors are active transporters, requiring cytoplasmic membrane energy to transport their substrates against a concentration gradient.[4] A number of members from all three OM protein classes have been studied in great detail, and a wealth of genetic, biochemical and structural information is available for them. However, all these proteins transport hydrophilic substrates; until recently, virtually nothing was known about how hydrophobic molecules cross the OM.
Interestingly, small hydrophobic molecules such as LCFAs do not utilize porins to traverse the OM. What is the reason for the inability of porins to transport hydrophobic substrates efficiently? X-ray crystallographic studies of porins such as OmpF have provided a likely answer to this question. A number of charged and polar residues surround the central, water-filled constriction of the channel, through which all substrates must pass. On one side of the constriction the residues are positively charged, while on the opposite side of the constriction negatively charged and electronegative residues dominate (Figure 1). This configuration likely creates a strong electric field across the constriction and will strongly orient the water molecules inside the pore, making it energetically unfavorable for a hydrophobic molecule to enter and traverse the pore.[5] Thus, the uptake of hydrophobic molecules across the OM requires specialized channels.
Figure 1.

Backbone representation of a monomer of the E. coli porin OmpF viewed from the extracellular side, showing the asymmetric distribution of charged residues at the water-filled constriction of the pore, shown as a green ellipse (blue, positively charged residues; red, negatively charged residues). The water molecules within the constriction are likely to be highly ordered, making passage of hydrophobic molecules energetically unfavorable.
3. The FadL family of OM channels
FadL channels are so far the only known OM channels with an established role in the uptake of hydrophobic compounds.[6] They are monomeric, substrate-specific channels that are widespread in Gram-negative bacteria. E. coli FadL, the archetype of the family, was first described over thirty years ago as a protein that confers the ability of E. coli to grow on LCFAs (> C12) as a sole carbon source.[7] A fadL knockout strain is, however, able to grow on medium-chain fatty acids, indicating that these more polar compounds can enter E. coli via other ways, presumably via porins. Subsequent work on bacterial fatty acid uptake, mainly from the group of Black[8,9], established FadL as an OM protein that works in concert with an inner membrane-associated fatty-acyl CoA synthetase (FadD) to drive LCFA transport into the cytoplasm, a process termed vectorial acylation (Scheme 3). Fadl-mediated uptake of LCFAs across the OM occurs via diffusion and does not require an external energy source. It does, however, depend on the presence of a sink inside the cytoplasm, e.g. in the absence of FadD no measurable transport of LCFAs occurs across the OM. The generated long chain fatty acyl-CoAs in the cytoplasm are the effector molecules that bind to the transcriptional regulator FadR, resulting in derepression of the fatty acid degradative genes, allowing the cell to utilize the fatty acids for metabolism (β-oxidation) and providing the driving force for transport.[8,9] It is of note that for this well-studied system, no inner membrane LCFA transport protein has been identified, which is consistent with the notion that LCFAs cross regular lipid bilayers spontaneously (Scheme 3).
Scheme 3.

Representation of LCFA transport in E. coli. LCFAs require a specialized OM channel (FadL) for uptake. Diffusion across the periplasm and inner membrane passage are likely to be spontaneous. FadD is the membrane-bound fatty acyl CoA synthetase, which activates the fatty acids so that they can be metabolized, providing the driving force for LCFA uptake.
4. The structure of E. coli FadL
The first X-ray crystal structures of E. coli FadL were solved about six years ago[10] and showed a 14-stranded β-barrel with two unique features (Figure 2). First, a small globular N-terminal domain (termed hatch or plug) completely blocks the lumen of the barrel, so that there is no channel connecting the extracellular milieu with the periplasmic space. Second, a segment of one of the β-strands (S3) does not form hydrogen bonds with its neighbor strands. This is due to the presence of a kink in the S3 strand, which interacts with the N-terminus of the hatch within the lumen of the barrel. A crucially important consequence of the presence of the kink is that there is a hole in the wall of the barrel (Figure 2). Besides these two unusual features of FadL, several detergent molecules (used for purification and crystallization of the protein) are bound at two hydrophobic binding pockets in the structure.[10] This is not surprising since LCFAs are detergents themselves; therefore the observed detergent molecules in FadL provide information about substrate binding.
Figure 2.
X-ray crystal structure of E. coli FadL. A) and B) Backbone representation of FadL viewed from the plane of the membrane, with B) a cut-away view 90 degrees rotated relative to A). The hatch domain is shown in purple, and the S3 kink, forming a hole in the side of the barrel, is shown in red. A bound detergent molecule, indicating where substrate would bind, is shown in green. The boundary of the hydrophobic core of the OM is shown as horizontal lines. C) Surface-filling view from the periplasmic space, showing that the hatch domain (purple) plugs the barrel and that there is no channel connecting the extracellular environment with the periplasmic space. D) Surface view of strand S2–S4, showing the lateral opening in the barrel wall created by the kink in strand S3 (red). For a clearer view of the lateral opening the background is shown in black.
5. Transport mechanism
Polar molecules are transported across biomembranes in a direction transversal to the membrane. Such a mechanism (termed "classical transport model") is possible for FadL as well. However, since the hatch plugs the barrel, a classical transport mechanism would require spontaneous conformational changes in the hatch domain to form a channel for substrate diffusion into the periplasmic space (Scheme 4). In addition to a classical transport model a second, alternative transport mechanism can be envisioned for FadL. Here, the substrates would diffuse from the lumen of the barrel laterally into the OM, via the opening in the barrel wall created by the kink in strand S3. The diameter of the opening is ~ 8 × 10 Å (atom-center to atom-center distance), and is therefore large enough to allow passage of a LCFA molecule. In the "lateral diffusion" model, the hatch would not move or undergo conformational changes. Instead, the hatch would function like a plug, preventing diffusion of the substrate directly into the periplasmic space (Scheme 4).
Scheme 4.

Two possible mechanisms of transport of hydrophobic molecules across the OM. The classical transport mechanism requires spontaneous conformational changes in the hatch domain (purple) to create a channel for substrate diffusion directly into the periplasmic space. The alternative, lateral diffusion model, is based on the presence of an opening in the barrel wall. According to this mechanism, substrates diffuse laterally from the lumen of the barrel into the OM via the lateral opening. See text for details.
To distinguish between the two possible transport models, a number of E. coli FadL mutants were generated within the kink and the hatch.[11] For the kink, two mutants were designed that would be expected to close the lateral opening, or at least would make it much smaller. In one of these mutants, A77E/S100R, two residues lining the lateral opening with small side chains (Ala, Ser) were replaced with residues possessing long side chains of opposite charge (Glu, Arg) that would interact with each other and therefore close the lateral opening. The functionality of the mutants was tested by in vivo LCFA uptake assays in E. coli, which demonstrated that both the lateral opening mutants did not transport LCFAs.[11] In addition, these mutants did not allow growth of E. coli on palmitic acid minimal medium plates. The functional assays were complemented with X-ray crystal structures of the lateral opening mutants, which demonstrated that the lateral opening in the mutants was much smaller (~ 4 × 6 Å in the A77E/S100R mutant), with the rest of the structure being identical to wild-type FadL. For the hatch, mutants were made within the absolutely conserved NPA sequence, with the rationale that modification of this region might induce conformational changes within the hatch. In agreement with the lateral diffusion model and arguing against a classical transport model, all NPA mutants had substantial transport activity.[11] Moreover, the NPA mutants had very similar structures compared to that of wild-type, i.e. no channels were observed through the hatch. Together the data demonstrated that constriction of the lateral opening is sufficient to block substrate transport, showing that uptake of hydrophobic molecules across the OM occurs by lateral diffusion through the opening in the barrel wall.[11] The lateral opening is conserved in FadL channels[11,12] and is very likely a stable structural feature. The position of the lateral opening is approximately at the interface region of the outer leaflet of the OM (Figure 2). This location makes sense, since it provides favorable environments for both the LCFA head group and the acyl chain upon emerging from the lateral opening. Since the substrate is deposited within the outer leaflet of the OM, flip-flop[1] to the inner leaflet will have to occur before the substrate can diffuse into the periplasmic space. The hatch domain is a static structure, preventing diffusion of the substrates directly into the periplasmic space, and should instead be designated as a "plug".
Why does uptake of hydrophobic molecules occur by lateral diffusion into the OM and not by a classical mechanism directly into the periplasmic space? The answer lies probably in the capacity of the OM to function as an efficient, large-capacity sink for hydrophobic molecules, driving transport forwards. From the OM, transport will be driven forward via desorption of the LCFA into the periplasmic space by simple mass action. Although desorption from the membrane in liposomes has been shown to become slower with longer acyl chains, it is still fast due to the high concentrations of LCFAs within the membrane.[1,2]
6. Transport via lateral diffusion through protein channel walls: a common theme?
It is likely that lateral diffusion does not only occur in channels belonging to the FadL family (Scheme 5). Likely candidates for channels that may operate via a similar mechanism as FadL are members of the OmpW family, which form small (8-stranded) β-barrels that are, like FadL channels, widespread in Gram-negative bacteria. Although functional data are scarce for this family, recent evidence suggests that they may be involved in the transport of small hydrophobic molecules across the OM.[13,14] Importantly, two X-ray crystal structures have been solved for OmpW members, and they both show a lateral opening in the barrel wall at a similar position as those in FadL channels.[15] OmpW channels do not have a plug; the 8-stranded barrels are sufficiently narrow so that interactions between residues located on opposite sides of the barrel close the barrel on the periplasmic side. Another example of a group of channels employing lateral diffusion may be proteins from COG4313, a group of uncharacterized outer membrane proteins that may be involved in uptake of small aromatic compounds. One family member was very recently implicated in the uptake of polychlorophenols.[16] No structural information is available for any COG4313 protein, but sequence alignments suggest that they are distinct from both OmpW and FadL channels.
Scheme 5.
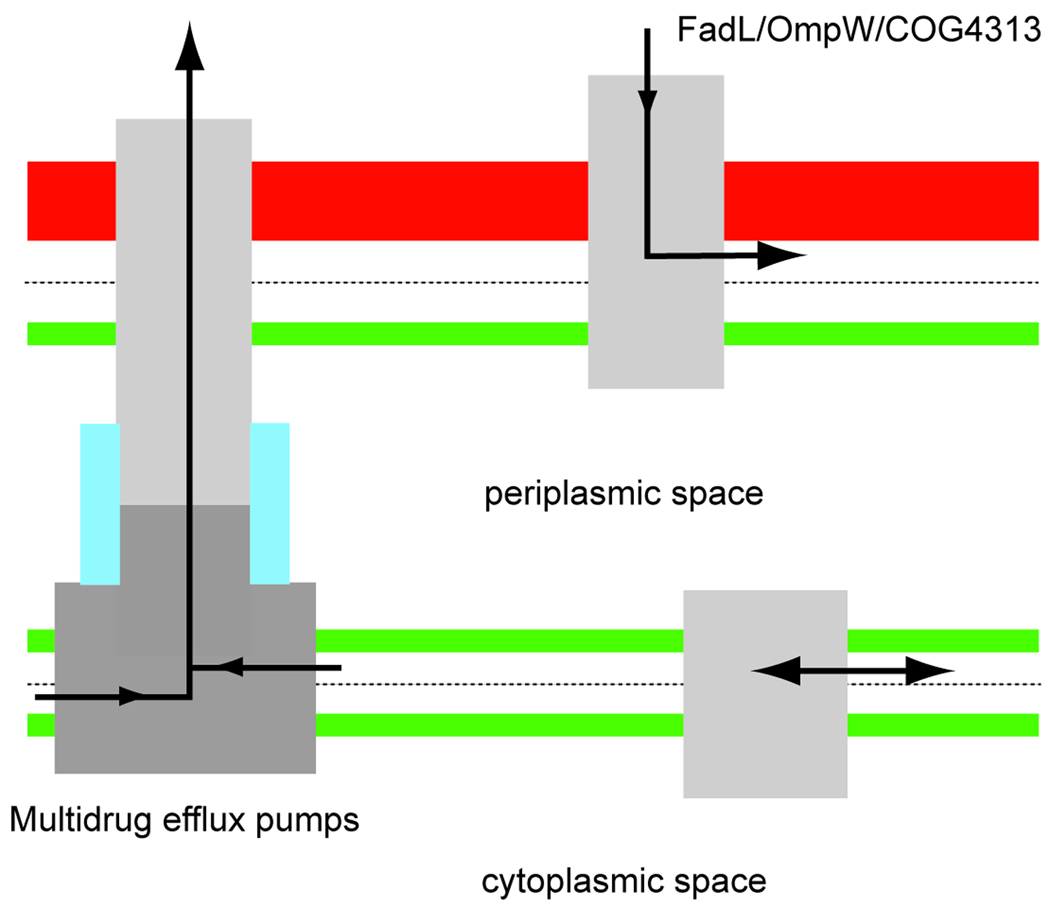
Lateral diffusion may be a widespread phenomenon in Nature for membrane proteins that transport and interact with hydrophobic molecules. See text for details.
Since OM transport of the above mentioned proteins is diffusion driven, it is conceivable that channels such as FadL might be used for the efflux of hydrophobic compounds from the outer membrane into the extracellular environment. However, no experimental evidence is yet available to support such a notion, and it should be noted that such efflux could only occur if the external concentration of the hydrophobic compound is lower than the intracellular one.
Perhaps surprisingly, there are also α-helical inner membrane proteins that use lateral diffusion as part of their transport mechanism. These are the inner membrane components of the RND (resistance nodulation division) multidrug efflux pumps, exemplified by the E. coli AcrB/AcrA/TolC system.[17,18] Here, hydrophobic drugs diffuse laterally from the inner membrane into large chambers within AcrB, upon which they are pumped in a single step across the periplasm and OM, out of the cell (Scheme 5). This is an active process, utilizing the proton gradient across the inner membrane as a source of energy. Thus, although the lateral diffusion step for multidrug efflux pumps occurs within the inner membrane, the required result is the same as for FadL channels, i.e. passage of the polar part of the LPS layer of the OM. The occurrence of lateral diffusion as part of transport processes may be limited to Gram-negative bacteria, since the cell wall of Gram-positive bacteria and eukaryotic cell membranes do not pose barriers for the passage of hydrophobic molecules. However, lateral diffusion through protein channel walls may be widespread in Nature for the many proteins that interact with and modify hydrophobic substrates.
Acknowledgements
The research on the uptake of hydrophobic molecules across the OM in the author's laboratory is supported by a grant from the NIH (NIGMS grant 5R01GM074824).
References
- 1.Hamilton JA. Prostaglandins Leukot Essent Fatty Acids. 2007;77:355–361. doi: 10.1016/j.plefa.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Simard JR, Pillai BK, Hamilton JA. Biochemistry. 2008;47:9081–9089. doi: 10.1021/bi800697q. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson AD, Deisenhofer J. Cell. 2004;116:15–24. doi: 10.1016/s0092-8674(03)01030-4. [DOI] [PubMed] [Google Scholar]
- 5.Schulz GE. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 6.van den Berg B. Curr Opin Struct Biol. 2005;15:401–407. doi: 10.1016/j.sbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Nunn WD, Simons RW. Proc Natl Acad Sci U S A. 1978;75:3377–3381. doi: 10.1073/pnas.75.7.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black PN, DiRusso CC. Microbiol Mol Biol Rev. 2003;67:454–472. doi: 10.1128/MMBR.67.3.454-472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black PN, Said B, Ghosn CR, Beach JV, Nunn WD WD. J Biol Chem. 1987;262:1412–1419. [PubMed] [Google Scholar]
- 10.van den Berg B, Black PN, Clemons WM, Jr, Rapoport TA. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]
- 11.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Nature. 2009;458:367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hearn EM, Patel DR, van den Berg B. Proc Natl Acad Sci U S A. 2008;105:8601–8606. doi: 10.1073/pnas.0801264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neher TM, Lueking DR. Can J Microbiol. 2009;55:553–563. doi: 10.1139/w09-002. [DOI] [PubMed] [Google Scholar]
- 14.van Beilen JB, Eggink G, Enequist H, Bos R, Witholt B. Mol Microbiol. 1992;6:3121–3136. doi: 10.1111/j.1365-2958.1992.tb01769.x. [DOI] [PubMed] [Google Scholar]
- 15.Hong H, Patel DR, Tamm LK, van den Berg B. J Biol Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- 16.Belchik SM, Schaeffer SM, Hasenoehrl S, Xun L. Biodegradation. 2009 Nov 24; doi: 10.1007/s10532-009-9313-8. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pos KM. Biochim Biophys Acta. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 18.Blair JM, Piddock LJ. Curr Opin Microbiol. 2009;12:512–519. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]



