Abstract
Hepatocellular carcinoma often develops in the setting of abnormal hepatocyte growth associated with chronic hepatitis and liver cirrhosis. Transforming growth factor-βs (TGF-βs) are multifunctional cytokines pivotal in the regulation of hepatic cell growth, differentiation, migration, extracellular matrix production, stem cell homeostasis and hepatocarcinogenesis. However, the mechanisms by which TGF-βs influence hepatic cell functions remain incompletely defined. We report herein that TGF-β regulates the growth of primary and transformed hepatocytes through concurrent activation of Smad and phosphorylation of cPLA2α, a rate-limiting key enzyme that releases arachidonic acid for production of bioactive eicosanoids. The interplays between TGF-β and cPLA2α signaling pathways were examined in rat primary hepatocytes, human hepatocellular carcinoma cells and hepatocytes isolated from the newly developed cPLA2α transgenic mice. Our data show that cPLA2α activates PPAR-γ and thus counteracts Smad2/3-mediated inhibition of cell growth. Therefore, regulation of TGF-β signaling by cPLA2α and PPAR-γ may represent an important mechanism for control of hepatic cell growth and hepatocarcinogenesis.
Keywords: Transforming growth factor-β, cytosolic phospholipase A2α, peroxisome proliferator activated receptor-γ, hepatocyte, liver
INTRODUCTION
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that plays an important role in the regulation of cell proliferation, differentiation, migration, apoptosis, extracellular matrix production, angiogenesis, and neoplasia(1-4). The actions initiated by TGF-β are complex and often vary depending on individual cell or tissue types and the activation status of other intracellular signaling pathways. In the liver, TGF-β is well known to regulate stellae cell activation/liver fibrosis, hepatocyte proliferation/apoptosis and hepatocarcinogenesis.
There are three mammalian TGF-β isoforms, TGF-β1, TGF-β2, and TGF-β3, all of which signal through a heteromeric complex of type I and type II TGF-β receptors(1, 5). Binding of ligand to the type II receptor results in the recruitment and activation of 1 of 2 type I receptors. The activated type I receptor phosphorylates Smad2 and Smad3. The phosphorylated Smad2/3 then associate with Smad4 and translocate to the nucleus. The mitoinhibition by TGF-β is predominantly mediated through activation of the Smad pathway. In addition, the activated TGFβRII and TGFβRI receptor complex can also signal independently of Smads, via extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK), p38 mitogen-activated protein kinase (MAPK), phosphatidylinositol-3 kinase (PI3K), and Rho GTPases(1, 2, 6). Recent studies have revealed an intriguing link between TGF-β and IL-6/Stat3 signaling pathways in hepatocarcinogenesis(7, 8). However, it remains unclear whether TGF-β may also modulate hepatocarcinogenesis through interaction with other growth-regulating signaling pathways. In this study we presented novel evidence that TGF-β regulates the growth of primary and transformed hepatocytes through concurrent activation of Smad-mediated gene transcription and phosphorylation of cPLA2α.
Prostaglandin (PG) signaling is implicated in the growth control of various human cells and cancers(9-14). PG biosynthesis is tightly controlled by a series of enzymes including the group IVα cytosolic phospholipase A2 (cPLA2α) that selectively cleaves arachidonic acid (AA) from membrane phospholipids, and cyclooxygenase-2 (COX-2) that converts AA substrate to PGs(9-14). This signaling cascade is active in various human cancers including hepatocellular carcinoma and promotes tumor growth by enhancing tumor cell proliferation, survival, invasion or angiogenesis(14-16). Whereas PGs regulate cell functions through activation of specific G protein-coupled receptors on plasma membrane, studies from our lab have shown that the cPLA2α-derived arachidonic acid can also regulate cellular functions through activation of nuclear receptors including peroxisome proliferator-activated receptor-γ (PPAR-γ)(17, 18).
This study describes the interaction between TGF-β and prostaglandin signaling pathways in primary and transformed hepatocytes. Our data show that TGF-β phosphorylates and activates cPLA2α and the cPLA2α-derived arachidonic acid subsequently activates PPAR-γ, leading to inhibition of Smad2/3. This phenomenon is further verified in hepatocytes isolated from the newly developed transgenic mice with targeted overexpression of cPLA2α in the liver. Our findings suggest that the level and activation status of cPLA2α/PPAR-γ in hepatic cells may represent a key factor that determines the cellular response to TGF-β and modulates hepatocarcinogenesis.
MATERIALS AND METHODS
Materials
Minimum essential medium with Earle's salts (EMEM), fetal bovine serum, glutamine, antibiotics, the Lipofectamine plus™ reagent and Lipofectamine™ 2000 reagent were purchased from Invitrogen (Carlsbad, CA). Human TGF-β1 (transforming growth factor beta 1) was purchased from R&D Systems, Inc. (Minneapolis, MN). Arachidonic acid(AA), prostaglandin E2(PGE2) were purchased from Calbiochem (San Diego, CA). The PPAR-γ agonists, ciglitazone and piglitazone, were purchased from Cayman Chemical (Ann Arbor, MI). The cPLA2α specific inhibitor pyrrolidine derivative, the COX2 inhibitor NS398, the p38 MAPK inhibitor SB203580 and the p42/44 MARK inhibitor PD98059 were purchased from Calbiochem (San Diego, CA). The antibodies against human cPLA2α, PPAR-γ, TGFβRI (transforming growth factor β receptor I), TGFβRII (transforming growth factor β receptor II), PAI-1 (the type 1 plasminogen activator inhibitor) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The antibodies against phospho-cPLA2α (Ser505), phospho-p38 MARP (Thr180/Tyr182), phospho-p42/44 MARP (Thr202/Tyr204), phospho-Smad2 (Ser465/467), phospho-Smad3 (Ser423/425), p38 MARP, p42/44 MARP, Smad2, Smad3, Smad4 were purchased from Cell Signaling (Berverly, MA). The antibody against β-actin was purchased from Sigma (St Louis, MO). The antibody against GAPDH was purchased from Ambion (Austin, TA). Amersham ECL Plus western blotting detection reagents were purchased from GE Healthcare (Piscataway, NJ). The cell proliferation assay reagent WST-1 was purchased from Roche Molecular Biochemicals (Indianapolis, IN). The PPAR-γ expression plasmid and PPRE-Luc reporter vector were purchased from Addgene (Cambridge, MA). The siRNAs for cPLA2α, PPAR-γ, Smad2 or Smad3 were purchased from Dharmacon (Chicago, IL).
Isolation of primary hepatocytes
Primary hepatocytes were isolated from the cPLA2α transgenic mice, matched wild type mice and Fisher 344 rats (Harlan, Indianapolis, IN) using the modified two-step collagenase perfusion technique as previously described (19, 20). Freshly isolated hepatocytes of >90% viability, as assessed by trypan blue exclusion, were placed on rat-tail collagen I-coated culture plates at a density of either 3 to 4 × 106 cells/100-mm plastic dish for western blotting or 1X 105 cells/each well of 6-well plates for the measurement of DNA synthesis in Williams’ E medium supplemented with 5% calf serum. The cells were incubated at 37°C (5% CO2) and checked for adherence of monolayers after 2 to 4 hours. Once adhered, the cells were changed to serum-free Williams’ E medium for 2 hours, then subjected to the treatment as indicated in the text.
Hepatocyte DNA Synthesis
The primary hepatocyte cultures were treated with either different concentrations of TGFβ1 or PGE2 as indicated in the text. To determine in vitro DNA synthesis, 1 μCi [3H]thymidine (PerkinElmer, Boston, MA) was added to each well of 6-well plates. After overnight incubation, the hepatocytes were harvested and [3H]-thymidine incorporation was measured using a scintillation counter.
Cell Growth Assay
Cell growth was determined using the cell proliferation reagent WST-1, a tetrazolium salt that is cleaved by mitochondrial dehydrogenases in viable cells. Briefly, 100 μl of cell suspension (containing 0.5–2 × 104 cells) were plated in each well of 96-well plates. After overnight culture to allow reattachment, the cells then were treated with specific reagents such as different concentrations of TGFβ1 in serum-free medium for indicated time points. At the end of each treatment, the cell proliferation reagent WST-1 (10 μl) was added to each well, and the cells were incubated at 37°C for 0.5–5 h. A450 nm was measured using an automatic ELISA plate reader.
Cell Culture and Transient Transfection
Three different human hepatocellular carcinoma cell lines (Hep3B, HepG2 and Huh7) were cultured according to our previously described methods (18, 21). For transient transfection assays, the cells with eighty percent confluence were transfected with the cPLA2α expression plasmid (with MT-2 as control plasmid) or the PPARγ expression plasmid (with pcDNA as control plasmid) using Lipofectamine plus™ reagent. The cells with optimal overexpression of either cPLA2α or PPARγ were confirmed by immunoblotting and subsequently used for further experiments.
Luciferase Reporter Assay
The cells with eighty percent confluence were transiently transfected with either p3TP or PPRE reporter vector using Lipofectamine plus™ reagent. After transfection, the cells were treated with specific reagent such as PPARγ agonists Ciglitazone and Piglitazone in serum-free medium for 24 hours. The cell lysates were then obtained with 1X reporter lysis buffer (Promega). The luciferase activity was assayed in a Berthold AutoLumat LB 953 luminometer (Nashua, NH) by using the luciferase assay system from Promega. The relative luciferase activity was calculated after normalization of cellular proteins. All values are expressed as –fold induction relative to basal activity.
Phosphorylation of cPLA2α
Analysis for cPLA2α phosphorylation was performed as we described previously(22). Equal amounts of cell lysate were preincubated with 5 μg/ml mouse anti-human cPLA2α monoclonal antibody for 1 hour followed by addition of 20 μl of protein A/G-agarose (Santa Cruz Biotechnology) for overnight at 4 °C. The cell lysate preincubated with mouse IgG was used as the negative control. After three washes with the same hypotonic buffer, the pellet was used for immunoblotting using rabbit anti-phospho-cPLA2α (Ser505) antibody.
DNA-protein Binding
DNA-protein binding was performed by the biotinylated oligonucleotides precipitation assay as described previously with minor modification (23). Briefly, 1 μg 5’-biotinylated, double stranded oligonucleotides that corresponded to the Smad3 binding site from the PAI-1 promoter region are forward: 5’-CAACCTCAGCCAGACAAGGTTGTTGACACAAGAGAGCCCTCAGGGGCACAGAGAGAGTCTGGACACGTGGGGAGTCAGCCGTGTATCATCGGAGGCGGCCGGGC-3’; and reverse: 5’-GCCCGGCCGCCTCCGATGATACACGGCTGACTCCCCACGTGTCCAGACTCTCTCTGTGCCCCTGAGGGCTCTCTTGTGTCAACAACCTTGTCTGGCTGAGGTTG-3’ (synthesized by Sigma-Genosys in Woodland, Texas) were mixed with equal amount of the cell extract and 10 μg poly(dl-dC).poly(dl-dC) for overnight at 4°C. After precipitated with ImmunoPure streptavidin-agarose beads (Pierce, Rockford, IL) at 4°C for another 1 hour, the DNA-bound proteins were subjected to detect Smad3 by immunoblotting.
RNA Interference
The cells with fifty percent confluence were transfected with either cPLA2α siRNA or PPAR-γ siRNA or Smad2/3 siRNA, or a 21-nucleotide irrelevant RNA duplex as a control siRNA using Lipofectamine™ 2000. After transfection the cells were cultured in serum-free medium for 48 hours. Depletion of cPLA2α or PPAR-γ or Smad2/3 was confirmed by immunoblotting for further experiments.
AA Release and PGE2 Production
To measure arachidonic acid release, the cells with eighty percent confluence in 6-well plates were labeled with 0.5 μCi/ml [3H]-arachidonic acid (218 Ci/mmol) in serum-supplemented medium for 18 hours. After washing three times with serum-free medium, the cells were exposed to TGF-β1 in the absence or presence of the cPLA2α specific inhibitor pyrrolidine derivative, the p38 MAPK inhibitor SB203580, or the p42/44 MAPK inhibitor PD98059. At the end of each treatment, the media were collected and centrifuged to remove the suspending cells. 0.5 ml of media was used for scintillation counting to measure 3H activity.
To measure PGE2 production, the serum-starved cells with eighty percent confluence in 6-well plates were exposed to TGF-β1 in the absence or presence of the inhibitors of cPLA2α, COX-2, p38 MAPK, or p42/44 MAPK in the serum-free medium. At the end of each treatment, the spent medium was collected. A 100-μl centrifuged sample was analyzed for PGE2 production using the PGE2 enzyme immunoassay system (Amersham Biosciences) according to the manufacturer's protocol.
Generation of cPLA2α transgenic mice
Transgenic mice with targeted expression of cPLA2α in the hepatocytes were developed by using the well-established albumin promoter-enhancer driven vector. To construct the albumin promoter-cPLA2α transgene, a 2.8 kb human cPLA2α cDNA containing the entire coding region of human cPLA2α was inserted into the first exon of the human growth hormone gene controlled by the mouse albumin ehancer/promoter(24, 25). After the confirmation of its overexpression in vitro, this transgene was micro-injected into mouse zygotes (B6SJL/F1 eggs) at the transgenic core facility of the University of Pennsylvania according to our contract. The produced transgenic lines were brought back to the University of Pittsburgh animal facility for propagation. The transgenic lines were maintained by backcrossing to the C57Bl/6 wild type mice and the transgenic mice were identified with genotyping from the genomic DNA of tails. The cPLA2α transgenic mice develop normally with no significant liver inflammation or histological abnormality under normal housing conditions, although the cPLA2α transgenic mice show slightly higher body weight and liver weight compared to the wild type mice (body weight - 25.82+1.23 in cPLA2α-Tg vs 22.63+0.68 in WT; liver weight - 1.30+0.10 in cPLA2α-Tg vs 1.13+0.06 in WT). The animals were kept at 22°C under a 12-h light/dark cycle and received food and water ad libitum. The handling of mice and experimental procedures were conducted in accordance with experimental animal guidelines. The cPLA2α transgenic mice used in this study were derived from one transgenic line that was backcrossed to C57Bl/6 wild type mice for 10 consecutive generations.
Statistical Analysis
All values were expressed as the mean and standard deviation. The statistical significance of differences between the groups was analyzed with the homoscedastic Student's t-Test, and p<0.05 was considered to be statistically significant. Single and double asterisks represent p<0.05 and p< 0.01, respectively.
RESULTS
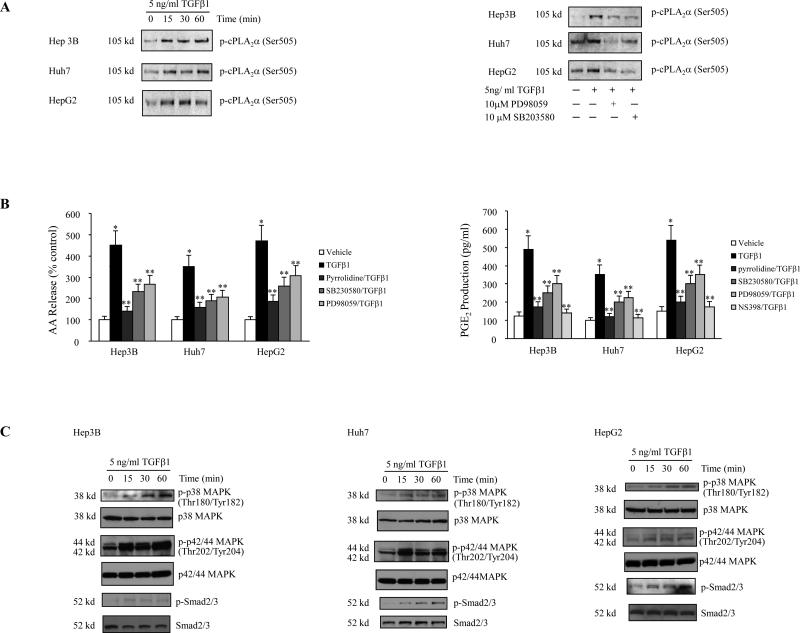
We examined the effect of TGF-β1 on cPLA2α phosphorylation and protein expression in three human hepatocellular carcinoma cell lines (Hep3B, Huh7 and HepG2). Treatment of these cells with TGF-β1 (5 ng/ml) induced a rapid phosphorylation of cPLA2α, occurring within 15 minutes (Figure 1A). In contrast, TGF-β1 treatment had no effect on the expression of cPLA2α and COX-2 proteins (Supplementary Figure 1). Consistent with its effect on cPLA2α phosphorylation, TGF-β1 treatment significantly increased AA release and PGE2 production in these cells (Figure 1B). These results show that TGF-β1 activates cPLA2α phosphorylation, increase AA release and PGE2 synthesis in these cells.
Figure 1. TGF-β1 activates arachidonic acid signaling cascade through cPLA2α phosphorylation in transformed human hepatocytes.
A. The effect of TGF-β1 on cPLA2α phosphorylation. (Left panel) TGF-β1 increases the phosphorylation of cPLA2α. Hep3B, Huh7 and HepG2 cells were treated with 5 ng/ml TGF-β1 for the indicated time periods. The cell lysates were then collected to determine the cPLA2α phosphorylation by immunoprecipitation and western blot. (Right panel) The p38 MAPK inhibitor SB203580 and the p42/44 MAPK inhibitor PD98059 inhibited TGF-β1-induced cPLA2α phosphorylation. Hep3B, Huh7 and HepG2 cells were treated with either 10 μM SB203580 or 10 μM PD98059 in serum-free medium for 2 hours prior to the stimulation with 5 ng/ml TGF-β1 for 15 minutes. The cell lysates were then collected to determine the cPLA2α phosphorylation by immunoprecipitation and western blot.
B. Arachidonice acid release and PGE2 production. (Left panel) The cPLA2α inhibitor pyrrolidine derivative, the p38 MAPK inhibitor SB203580, and the p42/44 MAPK inhibitor PD98059 inhibited TGF-β1-induced arachidonic acid (AA) release. Hep3B, Huh7 and HepG2 cells prelabeled with 0.5 μCi/ml [3H]-AA were treated with 5 ng/ml TGF-β1 for 60 minutes in the absence or presence of 2 μM pyrrolidine derivative, 10 μM SB203580 or 10 μM PD98059. The media were then collected for the measurement of AA release as indicated in Materials and Methods. The results are presented as mean ± SD of four experiments. (* p< 0.01, compared with control; ** p<0.05, compared with TGF-β1 treatment). (Right Panel) The cPLA2α inhibitor pyrrolidine derivative, the COX-2 inhibitor NS 398, the p38 MAPK inhibitor SB203580, and the p42/44 MAPK inhibitor PD98059 inhibited TGF-β1-induced PGE2 production. Hep3B, Huh7 and HepG2 cells were treated with 5 ng/ml TGF-β1 for 8 hours in the absence or presence of 2 μM pyrrolidine derivative, 25 μM NS398, 10 μM SB203580 or 10 μM PD98059 in serum-free medium. The media were then collected for the measurement of PGE2 production as indicated in Materials and Methods. The results are presented as mean ± SD of four experiments. (* p< 0.01, compared with control; ** p<0.05, compared with TGF-β1 treatment).
C. TGF-β1 activates p38 MAPK, p42/44 MAPK and Smad2/3. Hep3B, Huh7 and HepG2 cells were treated with 5 ng/ml TGF-β1 for the indicated time periods. The cell lysates were then collected to determine phosphorylated and total p38 MAPK, phosphorylated and total p42/44 MAPK, or phosphorylated and total Smad2/3 by western blot.
Since cPLA2α is phosphorylated by protein kinases including p38 MAPK and ERK1/2 (p44/42 MAPK), we also examined the effect of TGF-β1 on p38 MAPK and ERK1/2 activation in these cells. TGF-β1 treatment induced the phosphorylation of p38 MAPK and ERK1/2 as well as Smad2/3 (Figure 1C). These findings, along with the significant increase of Smad2/3 reporter activity by TGF-β1 (Fig 4C), indicate intact TGF-β-initiated signaling in these cells. The involvement of p38 MAPK and ERK1/2 in TGF-β1-induced cPLA2α phosphorylation is demonstrated by the fact that TGF-β1-induced cPLA2α phosphorylation in these cells was inhibited by the p38 MAPK inhibitor SB203580 and by the MEK1/2 inhibitor PD98059 (Figure 1A). Consistent with this, the TGF-β1-induced AA release and PGE2 production was also inhibited by SB203580, PD98059, and by the cPLA2α inhibitor pyrrolidine (Figure 1B). These findings demonstrate the role of p38 MAPK and ERK1/2-mediated cPLA2α activation in TGF-β1-induced AA release and PGE2 synthesis. In addition, the TGF-β1-induced PGE2 production was also inhibited by the selective COX-2 inhibitor NS-398 (Figure 1B), although the level of COX-2 expression was not altered (Supplementary Figure 1). Taken together, these findings suggest that TGF-β1 induces AA release for PGE2 production via p38 MAPK and ERK1/2-mediated cPLA2α phosphorylation.
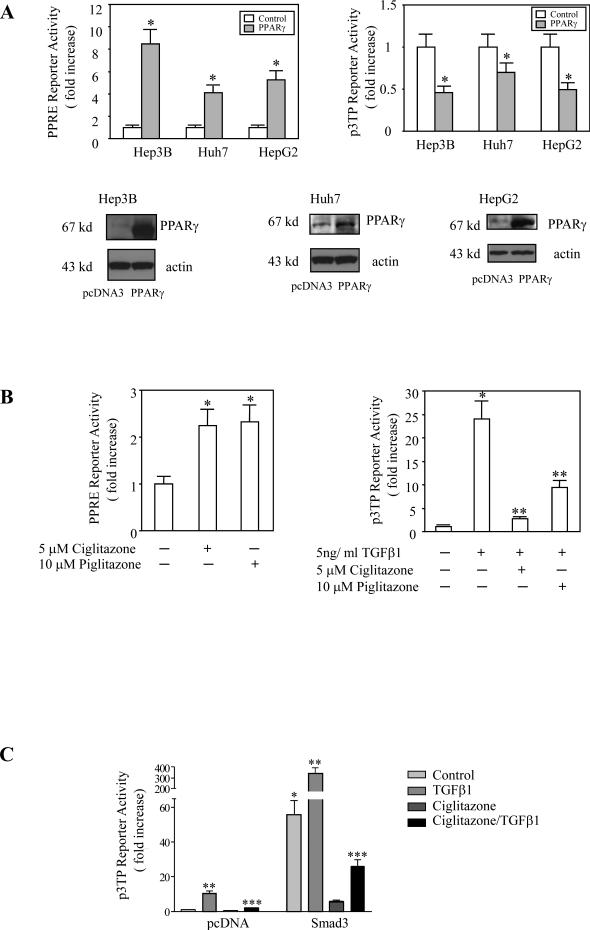
Figure 4. The effect of PPARγ overexpression and ligands on p3TP and PPRE reporter activities.
A. PPARγ overexpression. Hep3B, HepG2 and Huh 7 cells were transiently transfected with the PPARγ expression plasmid or pcDNA control plasmid with co-transfection of either PPRE-Luc reporter vector as shown in the left upper panel or the p3TP-Luc reporter vector as shown in the right upper panel. The cell lysates were obtained to determine the luciferase activity. The data are presented as mean ± S.D. of three independent experiments (* p<0.01 compared to corresponding vector control).
B. PPARγ ligands. (Left Panel) Hep3B cells were transiently transfected with PPRE-Luc reporter vector and then were treated with either 5 μM Ciglitazone or 10 μM Piglitazone in serum-free medium for 24 hours. The cell lysates were obtained to determine the luciferase activity. The data are presented as mean ± S.D. of three independent experiments (*p<0.01 compared with control). (Right Panel) Hep3B cells were transiently transfected with p3TP-Luc reporter vector. After transfection the cells were treated with 5 ng/ml TGF-β1 in the absence or presence of either Ciglitazone or Piglitazone in serum-free medium for 24 hours. The cell lysates were then obtained to determine the luciferase activity. The data are presented as mean ± S.D. of three independent experiments (*p<0.01 compared with no TGF-β1 treatment; **p<0.05 compared with TGF-β1 treatment alone).
C. The effect of ciglitazone on TGF-β1-induced p3TP reporter activity in cells with or without Smad3 overexpression. Hep3B cells were transiently transfected with either pcDNA control vector or Smad3 in pcDNA overexpression vector with co-transfection of the p3TP-Luc reporter vector. After transfection the cells were treated with 5 ng/ml TGF-β1 in the absence or presence of 5 μM Ciglitazone in serum-free medium for 24 hours; the cell lysates were then obtained to determine the luciferase activity. The data are presented as mean ± S.D. of three independent experiments. Overexpression of Smad 3 significantly increased the p3TP reporter activity compared with pcDNA control vector (*p<0.01). TGF-β1 significantly increased the p3TP reporter activity in pcDNA or Smad 3 transfected cells compared with each own control (**p<0.01). Ciglitazone significantly blocked TGF-β1-induced increase of p3TP reporter activity in cells transfected with either pcDNA or Smad3 in pcDNA (***p<0.01).
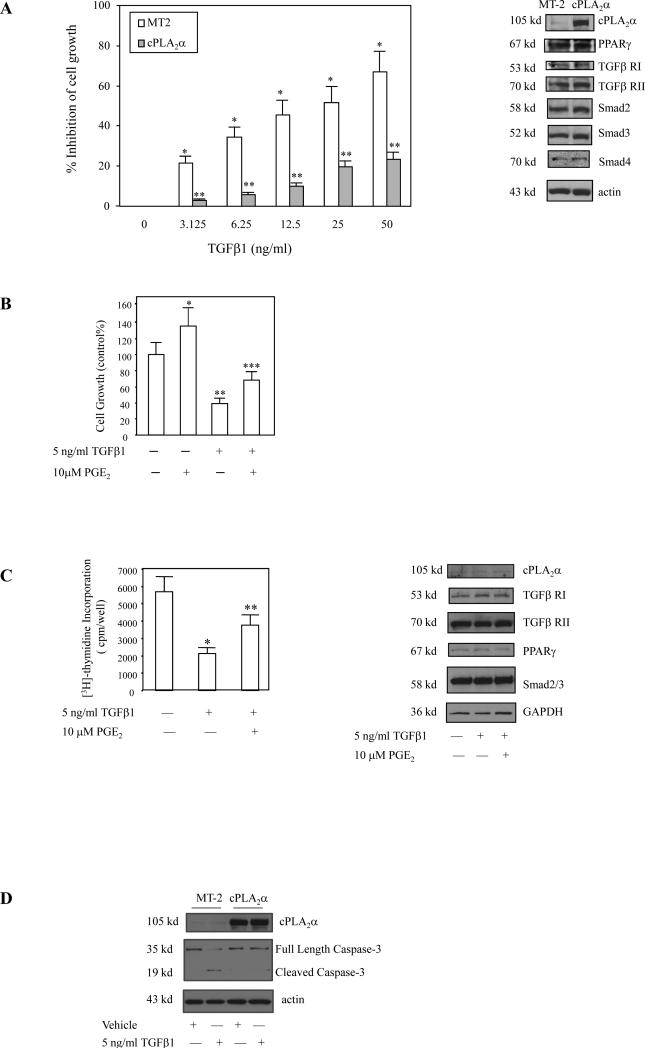
Further experiments were performed to determine whether cPLA2α overexpression or PGE2 treatment could prevent TGF-β1-induced inhibition of cell growth. As shown in Figure 2, overexpression of cPLA2α in Hep3B cells prevents TGF-β1-induced inhibition of growth; PGE2 treatment of Hep3B cells as well as rat primary hepatocytes also prevented TGF-β1-induced inhibition of cell growth. The observation that cPLA2α overexpression prevents TGF-β1-induced caspase-3 cleavage in Hep3B cells suggests the role of cPLA2α for prevention of TGF-β-induced apoptosis. These data indicate that cPLA2α signaling pathway is able to counteract the growth-inhibitory effect of TGF-β.
Figure 2. cPLA2α signaling prevents TGF-β1-induced inhibition of cell growth.
A. Overexpression of cPLA2α prevents TGF-β1-induced inhibition of Hep3B cell growth. Hep3B cells were transfected with either MT2 control vector or cPLA2α in MT2. After transfection, the cells were treated with different concentrations of TGF-β1 in serum-free medium for 48 hours. The cell growth was determined with WST-1 reagent; and the data are presented as mean ± S.D. of six independent experiments (* p<0.01 compared with MT2 control vector cells without TGF-β1 treatment; ** p<0.05 compared with MT2 control vector cells treated with the same concentration of TGF-β1). The Western blots in the right panel showed successful overexpression of cPLA2α in Hep3B cells transfected with the cPLA2α expression vector; the protein levels of PPARγ, TGFβRI, TGFβRII, Smad2, Smad3 and Smad4 were not altered.
B. PGE2 effect in Hep3B cells. Hep3B cells were treated with 5ng/ ml TGF-β1 in the absence or presence of 10μM PGE2 in serum-free medium for 48 hours. Cell growth was determined with the WST-1 reagent. The data are presented as mean ± S.D. of six independent experiments. PGE2 increased the growth of Hep3B cells (*p<0.05). TGF-β1 significantly inhibited the growth of Hep3B cells (**p<0.01); this effect was partially blocked by co-treatment with 10 μM PGE2 (*** p<0.05).
C. PGE2 effect in rat primary hepatocytes. Rat primary hepatocytes were treated with 5ng/ml TGF-β1 in the absence or presence of 10μM PGE2 in serum-free medium for 48 hours. Cell growth was determined with [3H]-thymidine incorporation assay (left panel). The data are presented as mean ± S.D. of three independent experiments (*p<0.01 compared with control; ** p<0.05 compared with TGF-β1 treatment). The Western blots in the right panel showed the protein levels of cPLA2α, PPAR-γ, TGFβRI, TGFβRII, Smad2/3 in rat primary hepatocytes.
D. cPLA2α overexpression prevents TGF-β1-induced caspase-3 activation. After transfection with the cPLA2α expression plasmid or the MT-2 control vector, the Hep3B cells were cultured in serum-free medium for 24 hours and then treated with either vehicle or 5 ng/ml TGFβ1 for 12 hours. The cell lysates were then obtained for western blotting analysis to determine Caspase-3 activation (β-actin was used as the loading control).
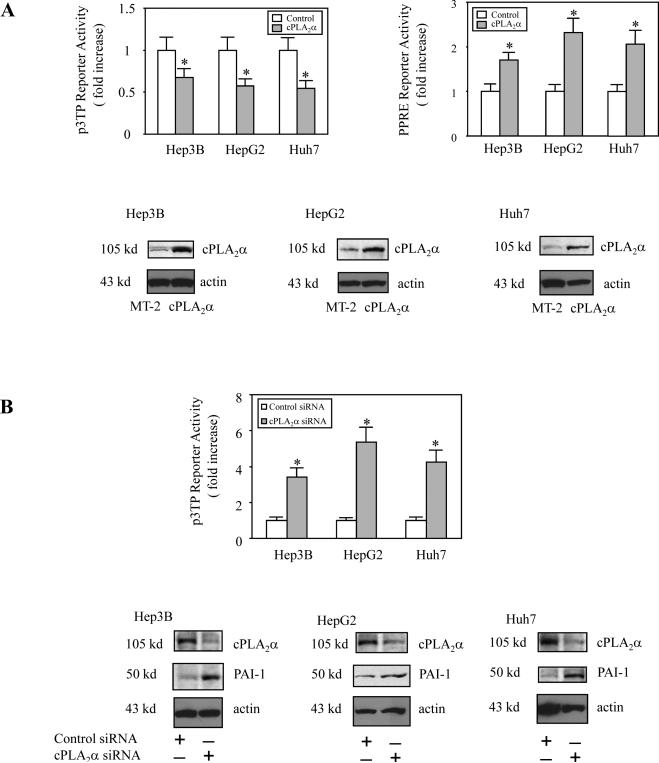
We next investigated whether the cPLA2α-mediated AA release might influence Smad transcriptional activity. Hep3B cells were transiently transfected with the cPLA2α expression plasmid or the control plasmid MT-2 with cotransfection of the p3TP-Lux reporter construct (containing Smad2/3-responsive element) and the cell lysates were obtained to determine the luciferase reporter activity. As shown in Figure 3A, overexpression of cPLA2α significantly inhibited Smad2/3 transcriptional activity. Accordingly, depletion of cPLA2α by siRNA significantly enhanced Smad2/3 transcriptional activity (Figure 3B). These findings reveal an important role of cPLA2α for modulation of Smad2/3 transcription activity.
Figure 3. The effect of cPLA2α on p3TP and PPRE reporter activities.
A. cPLA2α overexpression. Hep3B, HepG2 and Huh 7 cells were transiently transfected with the cPLA2α expression plasmid or MT2 control plasmid with co-transfection of either the p3TP-Luc reporter vector as shown in the left upper panel or the PPRE-Luc reporter vector as shown in the right upper panel. The cell lysates were obtained to determine the luciferase activity. The data are presented as mean ± S.D. of three independent experiments (* p<0.01 compared with corresponding control).
B. cPLA2α siRNA. Hep3B, HepG2 and Huh 7 cells were transiently transfected with either cPLA2α siRNA or control siRNA with co-transfection of p3TP-Luc reporter vector. The cell lysates were obtained to determine the luciferase activity. The data are presented as mean ± S.D. of three independent experiments (* p<0.01 compared to corresponding control siRNA). RNAi suppression of cPLA2α expression significantly increased the expression PAI-1, a Smad2/3 target gene.
PPARγ is a ligand-activated nuclear transcription factor regulating the expression of target genes by binding to specific peroxisome proliferator response elements (PPRE) or by interacting with other intracellular signaling molecules(26). The activity of PPAR-γ is regulated by several ligands, including thiazolidinediones, 15-deoxy-Δ12,14 prostaglandin J2, and arachidonic acid, among others. Consistent with our previous study showing that cPLA2α is able to activate PPAR-γ in other cells(17), cPLA2α overexpression was found to increase PPRE reporter (containing PPAR response element) activity in all three hepatocellular carcinoma cell lines used in this study (Hep3B, Huh7 and HepG2) (Figure 3A). Since PPAR-γ is known to bind and inhibit Smad3 in vitro(27) and cPLA2α is able to activate PPAR-γ, we postulated that cPLA2α might inhibit Smad3 through activation of PPAR-γ.
To document the direct effect of PPAR-γ on Smad activation, Hep3B, HepG2 and Huh7 cells were co-transfected with the PPAR-γ expression plasmid and the p3TP-Lux reporter construct containing the Smad2/3 response element. As shown in Figure 4A, overexpression of PPAR-γ partially inhibited Smad transcriptional activity in those cells. Accordingly, activation of PPAR-γ by its ligands (ciglitazone and piglitazone) significantly inhibited TGF-β1-induced Smad activation in Hep3 cells; this effect was observed with or without Smad3 overexpression (Figure 4B-C). In contrast, siRNA inhibition of PPAR-γ augmented TGF-β1-mediated Smad transcription (Figure 5A). In the transfection experiments with a reporter construct containing PPRE, we observed approximately 2 fold increase of PPRE reporter activity in Hep3B cells after PPAR-γ ligand treatment (ciglitazone and piglitazone) (Figure 4B), suggesting that the endogenous PPAR-γ protein in these cells is functional.
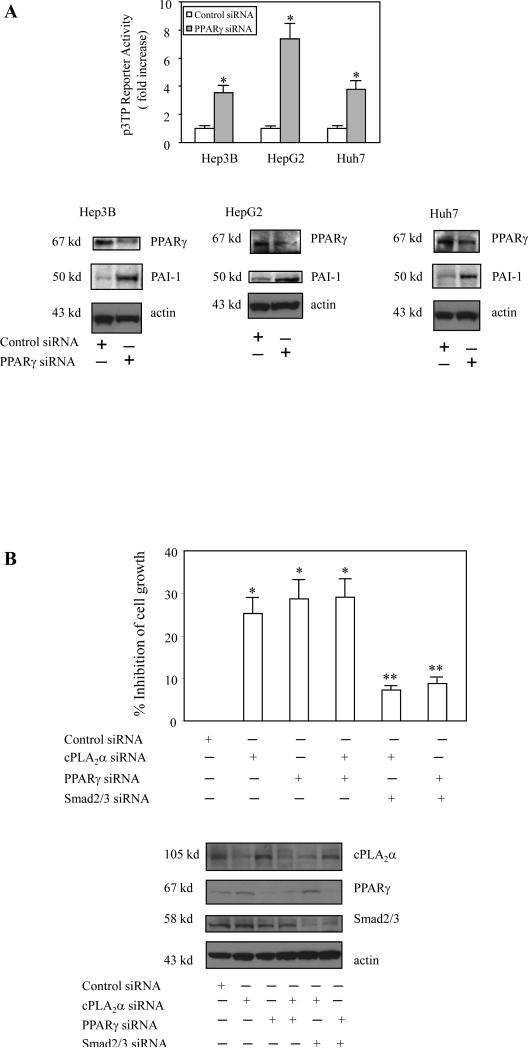
Figure 5. The effect of PPARγ depletion on p3TP reporter activity and cell growth.
A. PPARγ siRNA. Hep3B, HepG2 and Huh 7 cells were transiently transfected with either PPARγ siRNA or control siRNA with co-transfection of p3TP-Luc reporter vector. The cell lysates were obtained to determine the luciferase activity. The data are presented as mean ± S.D. of three independent experiments (* p<0.01 compared to control siRNA). RNAi suppression of PPARγ expression significantly increased the expression of the Smad2/3 target gene, PAI-1.
B. Depletion of cPLA2α and PPARγ inhibits cell growth – involvement of Smad2/3. Hep3B cells were transfected with cPLA2α siRNA or PPARγ siRNA, with or without Smad2/Smad3 siRNA. After transfection the cells were cultured in serum-free medium for 48 hours. The cell growth was determined with WST-1 reagent. The data are presented as mean ± S.D. of six independent experiments (* p<0.01 compared with control siRNA; **p<0.05 compared with cPLA2α siRNA or PPARγ siRNA). Western blots in the lower panel showed the successful depletion of cPLA2α, PPAR-γ or Smad2/3 in cells transfected with the corresponding siRNA.
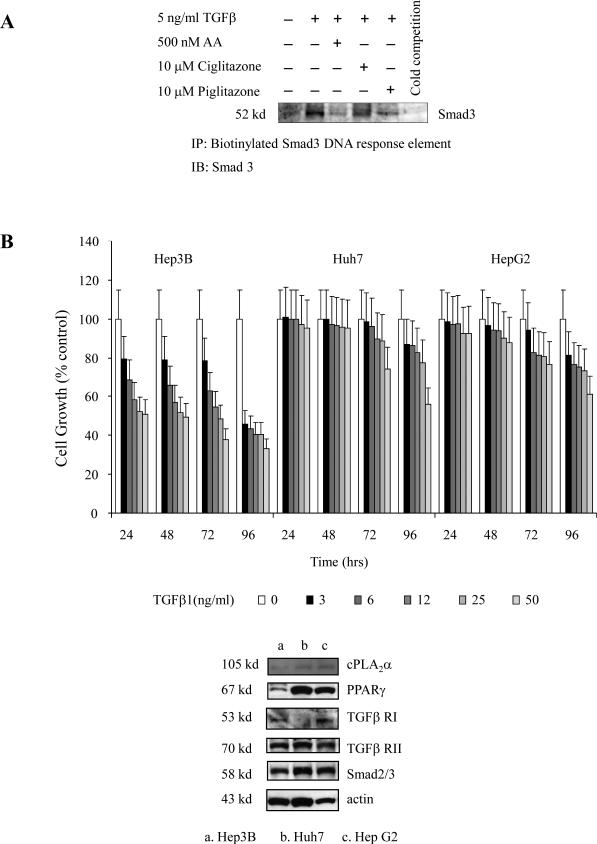
To further evaluate the role of cPLA2α and PPAR-γ in Smad activation and hepatic cell growth, additional experiments were performed to determine whether depletion of endogenous cPLA2α and PPAR-γ might inhibit cell growth via Smad2/3. As shown in Figure 5B, depletion of either cPLA2α or PPAR-γ significantly reduced cell growth and this effect was blocked by Smad2/Smad3 siRNA. These findings suggest the involvement of Smad2/3 in cPLA2α/PPAR-γ depletion-induced inhibition of cell growth. Further, the cPLA2α product, arachidonic acid, and the PPAR-γ ligands, ciglitazone and piglitazone, inhibited TGF-β1-induced binding of Smad 3 to its DNA response element (Figure 6A). Therefore, Smad3 is a downstream target of cPLA2α/PPAR-γ in hepatic cells.
Figure 6. AA and PPARγ ligands block TGF-β1-induced Smad 3 binding to its DNA response element. Comparison of cPLA2α and PPARγ expression in different cell lines.
A. Hep3B cells were treated with AA, Ciglitazone, Piglitazone in the absence or presence of TGF-β1 for 30 minutes. The binding of Smad3 to its DNA response element was analyzed by DNA-protein binding assay.
B. Hep3B, Huh7 and HepG2 cells were treated with different concentrations of TGF-β1 for different times as indicated. The cell growth was determined with WST-1 reagent; the data are presented as mean ± S.D. of six independent experiments. TGF-β1 inhibited the growth of Hep3B, but Huh7 and HepG2 cells. The Western blots in the lower panel showed the protein levels of cPLA2α, PPAR-γ, TGFβRI, TGFβRII and Smad2/3 in these cells.
We have found that cPLA2α activates PPAR-γ in all the three hepatic cell lines used in this study. However, these cell lines respond differently to TGF-β treatment – whereas TGF-β1 significantly inhibited the growth of Hep3B cells, it had minimal growth inhibitory effect in Huh7 or HepG2 cells under the same experimental condition (Figure 6B). The exact mechanism for such a differential effect among different cell lines is complex; but it is possible that this may partly relate to the low level of PPAR-γ expression in Hep3B cells (hence sensitive to TGF-β-induced inhibition of growth) and the high level of PPAR-γ in Huh7 and HepG2 cells (hence resistant to TGF-β-induced inhibition of growth).
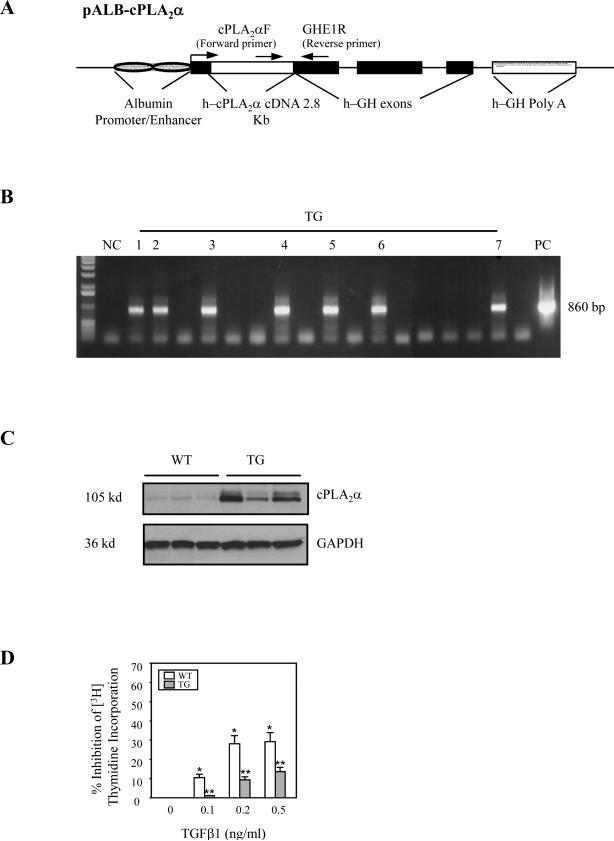
To further address the role of cPLA2α in TGF-β-induced hepatocyte growth regulation, we generated transgenic mice with targeted expression of the cPLA2α in the liver (Figure 7) and the produced animals were utilized to determine TGF-β-induced inhibition of hepatocyte growth. Primary hepatocytes were isolated from the cPLA2α transgenic or wild type mice and the cultured cells were treated with different concentrations of TGF-β1 in serum-free medium to determine [3H]-thymidine incorporation. As shown in Figure 7D, although TGF-β1 significantly inhibited the growth of hepatocytes from wild type mice, this effect was attenuated in cPLA2α overexpressed hepatocytes. Thus, overexpression of cPLA2α in hepatocytes renders the cells resistant to TGF-β1-induced inhibition of growth.
Figure 7. Overexpression of cPLA2α in hepatocytes prevents TGF-β1-induced mitoinhibition.
A. Schematic representation of the human cPLA2α transgene. Transgenic mice with targeted expression of cPLA2α in the hepatocytes were developed by using the well-established albumin promoter-enhancer driven vector. The 8 kb ALB-cPLA2α transgene consists of the 2.8 kb human cPLA2α cDNA (open box) inserted into the first exon of the human growth hormone gene (black boxes) controlled by the mouse albumin enhancer/promoter (cross hatched ovals), and possessing a human growth hormone polyadenylation site (checked box).
B. PCR analysis of tail genomic DNA for cPLA2α transgene. cPLA2α cDNA served as the positive control; the genomic DNA from wild type mouse served as a negative control. A 860 bp of cPLA2α transgene product was detected by PCR using specific primer pairs (forward primer: CPLA2αF: 5’-TGGCCAACATCAACTTCAGA-3’ and reverse primer: GHE1R: 5’TTACCTGCAGCCATTGCCGCTAGTGAG-3’) derived from pALB-cPLA2α transgene.
C. Western blotting analysis of protein levels in liver tissues from cPLA2α transgenic mice. Equal amounts of the liver tissue proteins from wild-type (WT) and cPLA2α transgenic mice (TG) littermates were subjected to SDS-PAGE and Western blot using anti-human antibody (with GAPDH as a loading control).
D. TGF-β1-mediated mitoinhibition of primary hepatocytes from cPLA2α transgenic and wild type mice. Primary hepatocytes isolated from the wild type or cPLA2α transgenic mice were treated with different concentrations of TGF-β1 in serum-free medium for 48 hours. Cell proliferation was determined with [3H]-thymidine incorporation assay. The data are presented as mean ± S.D. of three independent experiments (*p<0.01 compared wild type cells without TGF-β1 treatment; **p<0.05 compared with the same TGF-β1 treatment of wild type cells).
DISCUSSION
In the liver, TGF-β is well-known to inhibit hepatocyte proliferation and induce hepatocyte apoptosis(28-34). Quiescent liver usually contains only modest amounts of TGF-β but injury to the liver results in the production of TGF-β, most prominently by nonparenchymal cells, including hepatic stellate cells and Kupffer cells. TGF-β1 regulates hepatocyte growth by inducing cell cycle arrest or apoptosis, both in vitro and in vivo(28-34). In primary hepatocyte cultures, TGF-β inhibits DNA synthesis from normal and regenerating livers by blocking the transition from the G1 to the S phase of the cell cycle. After a two-thirds partial hepatectomy, TGF-β1 mRNA expression increases, and TGF-β is the ostensible inhibitory peptide for hepatocyte replication and liver regeneration(35). Intravenous administration of mature TGF-β to rats has been shown to reduce [3H]thymidine incorporation in hepatocytes after partial hepatectomy, although inhibition of hepatocyte DNA synthesis was transient since complete regeneration of the liver still occurred by 8 days(30). On the other hand, intravenous administration of adenoviral vector expressing active TGF-β1 potently inhibits liver regeneration in rats after standard two thirds partial hepatectomy, ultimately leading to animal death(36). Thus, high levels of expression of active TGF-β1 is able to effectively inhibit hepatocyte growth in vivo.
In consideration of the key role of TGF-β in the regulation of hepatocyte growth and liver fibrosis, it is not surprising that TGF-β is causatively involved in hepatocarcinogenesis. Given its antiproliferative and proapoptotic role in the liver, TGF-β1 could be expected to act as a tumor suppressor. Indeed, mice heterozygous for the deletion of TGF-β1 or TGFβRII were found to be more susceptible to DEN-induced hepatocarcinogenesis when compared to their wild-type littermates(37, 38) (mice homozygous for the deletion of TGF-β1 or TGFβRII are lethal). Enhanced hepatocarcinogenesis is also observed in transgenic mice overexpressing a dominant negative TGFβRII(39) or in mice heterozygous for deletion of the Smad adaptor protein, embryonic liver fodrin (ELF)(40). Consistent with these observations, forced expression of Smad3 in the liver has been shown to inhibit DEN-induced hepatocarcinogenesis(41). However, there is also evidence suggesting that TGF-β may promote hepatic carcinogenesis(42-44) and that late stage human liver cancers often show overexpression of TGF-β (45, 46). The opposing effects of TGF-β on liver cancer growth may be explained by the fact that TGF-βs function as tumor suppressors early in tumorigenesis when epithelial cell responsiveness to TGF-β is still relatively normal. It is possible that during multistage tumorigenesis, the mitoinhibitory effect of TGF-βs becomes lost, either through mutation of the TGF-β signaling molecules or by subversion of the normal signaling pathway due to activation of other molecules(1, 2, 47-49). Since mutation of TGF-β signaling molecules occurs only in a minority of human hepatocellular cancers and the prostaglandin signaling pathway is active in tumor cells(15, 50), we postulate that disruption of TGF-β-mediated inhibition of cell growth by prostaglandin cascade may be an important mechanism for regulation of cell growth and carcinogenesis.
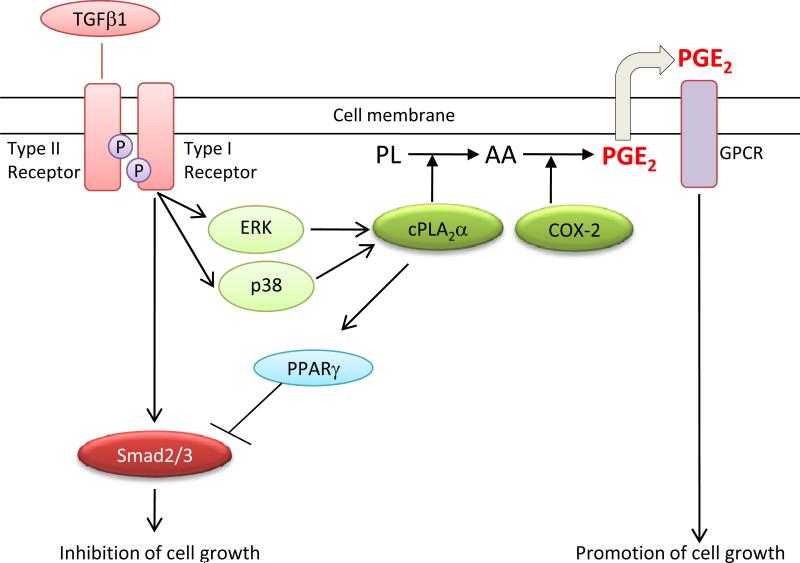
The current study shows that TGF-β regulates the growth of primary and transformed hepatocytes through concurrent activation of Smad-mediated gene transcription and phosphorylation of cPLA2α (as illustrated in Figure 8). Our findings suggest that the level and activation status of cPLA2α/PPAR-γ signaling in hepatic cells likely represents a key factor that determines the cellular response to TGF-β. It is possible that activation of cPLA2α/PPAR-γ signaling may in part explain the loss of responsiveness of neoplastic cells to the antiproliferative actions of TGF-β (due to suppression of Smad2/3 activity). Further studies are warranted to determine the exact role of this interaction in different stages of hepatocarcinogenesis.
Figure 8.
Schematic illustration of the interaction between cPLA2α and TGF-βsignaling pathways in hepatic cells.
Supplementary Material
Supplementary Methods
Immunoblotting. At the end of each indicated treatment, the cells scraped off the plates were centrifuged and washed twice with cold phosphate-buffered saline (PBS) containing 0.5 mM PMSF and 10μg/ml leupeptin. The cell pellet was sonicated in 5-fold volume of hypotonic buffer consisting of 50 mM HEPES pH 7.55, 1 mM EDTA, 1 mM DTT, protease inhibitor cocktail tablets (Roche Diagnostics GmbH). After sonication, the whole cell lysate was collected by centrifugation at the speed of 16,000 × g at 4°C for 10 minutes to remove cell debris and stored in aliquots at -20°C until use. The protein concentrations in the cell extracts were determined by the Bio-Rad protein assay (Bio-Rad, CA) using BSA as a standard. 30 μg of cellular protein was subjected to SDS-PAGE on either 4-20% or 10% Tris-glycine gels (Invitrogen, CA). The separated proteins were electrophoretically transferred onto the nitrocellulose membranes (BioRad, CA). Nonspecific binding was blocked with PBS-T (0.5% Tween 20 in PBS) containing 5% non-fat milk for 1 hr at room temperature. The membranes were then incubated overnight at 4°C with individual primary antibodies in PBS-T containing 1% non-fat milk at the dilutions specified by the manufacturers. Following three washes with PBS-T, the membranes were then incubated with the horseradish peroxidase-conjugated secondary antibodies at 1:10,000 dilution in PBS-T containing 1% non-fat milk for 1 hour at room temperature. The membranes were then washed 3 times with PBS-T and the protein bands were visualized with the ECL Western blotting detection system. The equal amount of cellular proteins was verified by either β-actin for HCC cells or GAPDH for primary hepatocytes as a loading control.
Supplementary Figure 1 Legend
TGF-β1 does not alter the expression of cPLA2α and COX-2 proteins. Hep3B, Huh7 and HepG2 cells were treated with 5 ng/ml TGF-β1 for 6-96 hours and the cell lysates were obtained to determine the protein levels of cPLA2α, COX2 and Smad 2/3 by western blot.
Acknowledgments
Supported by National Institutes of Health grants DK077776, CA106280, CA102325 and CA134568 (to T.W.) and CA137729 (to C.H.).
ABBREVIATIONS
- AA
arachidonic acid
- COX-2
cyclooxygenase-2
- cPLA2α
cytosolic phospholipase A2α
- MAPK
mitogen-activated protein kinase
- PG
prostaglandin
- PGE2
prostaglandin E2
- PPARγ
peroxisome proliferator activated receptor γ
- PPRE
peroxisome proliferator response element
- TGF-β
transforming growth factor-β
REFERENCES
- 1.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 2.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 3.Mishra L, Derynck R, Mishra B. Transforming growth factor-beta signaling in stem cells and cancer. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, Moses HL. Transforming growth factor beta: tumor suppressor or promoter? Are host immune cells the answer? Cancer Res. 2008;68:9107–9111. doi: 10.1158/0008-5472.CAN-08-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 7.Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC, Ressom HW, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci U S A. 2008;105:2445–2450. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin L, Amin R, Gallicano GI, Glasgow E, Jogunoori W, Jessup JM, Zasloff M, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28:961–972. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res. 2009;50(Suppl):S237–242. doi: 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaloske RH, Dennis EA. The phospholipase A2 superfamily and its group numbering system. Biochim Biophys Acta. 2006;1761:1246–1259. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh M, Tucker DE, Burchett SA, Leslie CC. Properties of the Group IV phospholipase A2 family. Prog Lipid Res. 2006;45:487–510. doi: 10.1016/j.plipres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Murakami M, Kudo I. Phospholipase A2. J Biochem (Tokyo) 2002;131:285–292. doi: 10.1093/oxfordjournals.jbchem.a003101. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick FA, Soberman R. Regulated formation of eicosanoids. J Clin Invest. 2001;107:1347–1351. doi: 10.1172/JCI13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1:11–21. doi: 10.1038/35094017. [DOI] [PubMed] [Google Scholar]
- 15.Wu T. Cyclooxygenase-2 in hepatocellular carcinoma. Cancer Treat Rev. 2006;32:28–44. doi: 10.1016/j.ctrv.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Wu T. Cyclooxygenase-2 and prostaglandin signaling in cholangiocarcinoma. Biochim Biophys Acta. 2005;1755:135–150. doi: 10.1016/j.bbcan.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Pawliczak R, Han C, Huang XL, Demetris AJ, Shelhamer JH, Wu T. 85-kDa cytosolic phospholipase A2 mediates peroxisome proliferator-activated receptor gamma activation in human lung epithelial cells. J Biol Chem. 2002;277:33153–33163. doi: 10.1074/jbc.M200246200. [DOI] [PubMed] [Google Scholar]
- 18.Xu L, Han C, Lim K, Wu T. Cross-talk between peroxisome proliferator-activated receptor delta and cytosolic phospholipase A(2)alpha/cyclooxygenase-2/prostaglandin E(2) signaling pathways in human hepatocellular carcinoma cells. Cancer Res. 2006;66:11859–11868. doi: 10.1158/0008-5472.CAN-06-1445. [DOI] [PubMed] [Google Scholar]
- 19.Seglen PO. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 20.Runge D, Runge DM, Jager D, Lubecki KA, Beer Stolz D, Karathanasis S, Kietzmann T, et al. Serum-free, long-term cultures of human hepatocytes: maintenance of cell morphology, transcription factors, and liver-specific functions. Biochem Biophys Res Commun. 2000;269:46–53. doi: 10.1006/bbrc.2000.2215. [DOI] [PubMed] [Google Scholar]
- 21.Han C, Leng J, Demetris AJ, Wu T. Cyclooxygenase-2 promotes human cholangiocarcinoma growth: evidence for cyclooxygenase-2-independent mechanism in celecoxib-mediated induction of p21waf1/cip1 and p27kip1 and cell cycle arrest. Cancer Res. 2004;64:1369–1376. doi: 10.1158/0008-5472.can-03-1086. [DOI] [PubMed] [Google Scholar]
- 22.Wu T, Han C, Lunz JG, 3rd, Michalopoulos G, Shelhamer JH, Demetris AJ. Involvement of 85-kd cytosolic phospholipase A(2) and cyclooxygenase-2 in the proliferation of human cholangiocarcinoma cells. Hepatology. 2002;36:363–373. doi: 10.1053/jhep.2002.34743. [DOI] [PubMed] [Google Scholar]
- 23.Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massague J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 24.Bell A, Chen Q, DeFrances MC, Michalopoulos GK, Zarnegar R. The five amino acid-deleted isoform of hepatocyte growth factor promotes carcinogenesis in transgenic mice. Oncogene. 1999;18:887–895. doi: 10.1038/sj.onc.1202379. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, DeFrances MC, Dai Y, Pediaditakis P, Johnson C, Bell A, Michalopoulos GK, et al. A Mechanism of Cell Survival. Sequestration of Fas by the HGF Receptor Met. Mol Cell. 2002;9:411–421. doi: 10.1016/s1097-2765(02)00439-2. [DOI] [PubMed] [Google Scholar]
- 26.Rosen ED, Spiegelman BM. Ppargamma : a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 27.Fu M, Zhang J, Zhu X, Myles DE, Willson TM, Liu X, Chen YE. Peroxisome proliferator-activated receptor gamma inhibits transforming growth factor beta-induced connective tissue growth factor expression in human aortic smooth muscle cells by interfering with Smad3. J Biol Chem. 2001;276:45888–45894. doi: 10.1074/jbc.M105490200. [DOI] [PubMed] [Google Scholar]
- 28.Carr BI, Hayashi I, Branum EL, Moses HL. Inhibition of DNA synthesis in rat hepatocytes by platelet-derived type beta transforming growth factor. Cancer Res. 1986;46:2330–2334. [PubMed] [Google Scholar]
- 29.Nakamura T, Tomita Y, Hirai R, Yamaoka K, Kaji K, Ichihara A. Inhibitory effect of transforming growth factor-beta on DNA synthesis of adult rat hepatocytes in primary culture. Biochem Biophys Res Commun. 1985;133:1042–1050. doi: 10.1016/0006-291x(85)91241-0. [DOI] [PubMed] [Google Scholar]
- 30.Russell WE, Coffey RJ, Jr., Ouellette AJ, Moses HL. Type beta transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc Natl Acad Sci U S A. 1988;85:5126–5130. doi: 10.1073/pnas.85.14.5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun L, Mead JE, Panzica M, Mikumo R, Bell GI, Fausto N. Transforming growth factor beta mRNA increases during liver regeneration: a possible paracrine mechanism of growth regulation. Proc Natl Acad Sci U S A. 1988;85:1539–1543. doi: 10.1073/pnas.85.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakowlew SB, Mead JE, Danielpour D, Wu J, Roberts AB, Fausto N. Transforming growth factor-beta (TGF-beta) isoforms in rat liver regeneration: messenger RNA expression and activation of latent TGF-beta. Cell Regul. 1991;2:535–548. doi: 10.1091/mbc.2.7.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bottinger EP, Factor VM, Tsang ML, Weatherbee JA, Kopp JB, Qian SW, Wakefield LM, et al. The recombinant proregion of transforming growth factor beta1 (latency-associated peptide) inhibits active transforming growth factor beta1 in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:5877–5882. doi: 10.1073/pnas.93.12.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero-Gallo J, Sozmen EG, Chytil A, Russell WE, Whitehead R, Parks WT, Holdren MS, et al. Inactivation of TGF-beta signaling in hepatocytes results in an increased proliferative response after partial hepatectomy. Oncogene. 2005;24:3028–3041. doi: 10.1038/sj.onc.1208475. [DOI] [PubMed] [Google Scholar]
- 35.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrum LW, Bird MA, Salcher O, Burchardt ER, Grisham JW, Brenner DA, Rippe RA, et al. Autocrine expression of activated transforming growth factor-beta(1) induces apoptosis in normal rat liver. Am J Physiol Gastrointest Liver Physiol. 2001;280:G139–148. doi: 10.1152/ajpgi.2001.280.1.G139. [DOI] [PubMed] [Google Scholar]
- 37.Tang B, Bottinger EP, Jakowlew SB, Bagnall KM, Mariano J, Anver MR, Letterio JJ, et al. Transforming growth factor-beta1 is a new form of tumor suppressor with true haploid insufficiency. Nat Med. 1998;4:802–807. doi: 10.1038/nm0798-802. [DOI] [PubMed] [Google Scholar]
- 38.Im YH, Kim HT, Kim IY, Factor VM, Hahm KB, Anzano M, Jang JJ, et al. Heterozygous mice for the transforming growth factor-beta type II receptor gene have increased susceptibility to hepatocellular carcinogenesis. Cancer Res. 2001;61:6665–6668. [PubMed] [Google Scholar]
- 39.Kanzler S, Meyer E, Lohse AW, Schirmacher P, Henninger J, Galle PR, Blessing M. Hepatocellular expression of a dominant-negative mutant TGF-beta type II receptor accelerates chemically induced hepatocarcinogenesis. Oncogene. 2001;20:5015–5024. doi: 10.1038/sj.onc.1204544. [DOI] [PubMed] [Google Scholar]
- 40.Kitisin K, Ganesan N, Tang Y, Jogunoori W, Volpe EA, Kim SS, Katuri V, et al. Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene. 2007 doi: 10.1038/sj.onc.1210513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang YA, Zhang GM, Feigenbaum L, Zhang YE. Smad3 reduces susceptibility to hepatocarcinoma by sensitizing hepatocytes to apoptosis through downregulation of Bcl-2. Cancer Cell. 2006;9:445–457. doi: 10.1016/j.ccr.2006.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Factor VM, Kao CY, Santoni-Rugiu E, Woitach JT, Jensen MR, Thorgeirsson SS. Constitutive expression of mature transforming growth factor beta1 in the liver accelerates hepatocarcinogenesis in transgenic mice. Cancer Res. 1997;57:2089–2095. [PubMed] [Google Scholar]
- 43.Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, Roberts AB, et al. Hepatic expression of mature transforming growth factor beta 1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci U S A. 1995;92:2572–2576. doi: 10.1073/pnas.92.7.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnur J, Nagy P, Sebestyen A, Schaff Z, Thorgeirsson SS. Chemical hepatocarcinogenesis in transgenic mice overexpressing mature TGF beta-1 in liver. Eur J Cancer. 1999;35:1842–1845. doi: 10.1016/s0959-8049(99)00224-5. [DOI] [PubMed] [Google Scholar]
- 45.Bedossa P, Peltier E, Terris B, Franco D, Poynard T. Transforming growth factor-beta 1 (TGF-beta 1) and TGF-beta 1 receptors in normal, cirrhotic, and neoplastic human livers. Hepatology. 1995;21:760–766. [PubMed] [Google Scholar]
- 46.Ito N, Kawata S, Tamura S, Takaishi K, Shirai Y, Kiso S, Yabuuchi I, et al. Elevated levels of transforming growth factor beta messenger RNA and its polypeptide in human hepatocellular carcinoma. Cancer Res. 1991;51:4080–4083. [PubMed] [Google Scholar]
- 47.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 48.Akhurst RJ, Derynck R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001;11:S44–51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 49.Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–29. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 50.Leng J, Han C, Demetris AJ, Michalopoulos GK, Wu T. Cyclooxygenase-2 promotes hepatocellular carcinoma cell growth through Akt activation: evidence for Akt inhibition in celecoxib-induced apoptosis. Hepatology. 2003;38:756–768. doi: 10.1053/jhep.2003.50380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Immunoblotting. At the end of each indicated treatment, the cells scraped off the plates were centrifuged and washed twice with cold phosphate-buffered saline (PBS) containing 0.5 mM PMSF and 10μg/ml leupeptin. The cell pellet was sonicated in 5-fold volume of hypotonic buffer consisting of 50 mM HEPES pH 7.55, 1 mM EDTA, 1 mM DTT, protease inhibitor cocktail tablets (Roche Diagnostics GmbH). After sonication, the whole cell lysate was collected by centrifugation at the speed of 16,000 × g at 4°C for 10 minutes to remove cell debris and stored in aliquots at -20°C until use. The protein concentrations in the cell extracts were determined by the Bio-Rad protein assay (Bio-Rad, CA) using BSA as a standard. 30 μg of cellular protein was subjected to SDS-PAGE on either 4-20% or 10% Tris-glycine gels (Invitrogen, CA). The separated proteins were electrophoretically transferred onto the nitrocellulose membranes (BioRad, CA). Nonspecific binding was blocked with PBS-T (0.5% Tween 20 in PBS) containing 5% non-fat milk for 1 hr at room temperature. The membranes were then incubated overnight at 4°C with individual primary antibodies in PBS-T containing 1% non-fat milk at the dilutions specified by the manufacturers. Following three washes with PBS-T, the membranes were then incubated with the horseradish peroxidase-conjugated secondary antibodies at 1:10,000 dilution in PBS-T containing 1% non-fat milk for 1 hour at room temperature. The membranes were then washed 3 times with PBS-T and the protein bands were visualized with the ECL Western blotting detection system. The equal amount of cellular proteins was verified by either β-actin for HCC cells or GAPDH for primary hepatocytes as a loading control.
Supplementary Figure 1 Legend
TGF-β1 does not alter the expression of cPLA2α and COX-2 proteins. Hep3B, Huh7 and HepG2 cells were treated with 5 ng/ml TGF-β1 for 6-96 hours and the cell lysates were obtained to determine the protein levels of cPLA2α, COX2 and Smad 2/3 by western blot.