Abstract
Significant variation in the course of autosomal dominant polycystic kidney disease ( ADPKD) within families suggests the presence of effect modifiers. Recent studies of the variation within families harboring PKD1 mutations indicate that genetic background may account for 32 to 42% of the variance in estimated GFR (eGFR) before ESRD and 43 to 78% of the variance in age at ESRD onset, but the genetic modifiers are unknown. Here, we conducted a high-throughput single-nucleotide polymorphism (SNP) genotyping association study of 173 biological candidate genes in 794 white patients from 227 families with PKD1. We analyzed two primary outcomes: (1) eGFR and (2) time to ESRD (renal survival). For both outcomes, we used multidimensional scaling to correct for population structure and generalized estimating equations to account for the relatedness among individuals within the same family. We found suggestive associations between each of 12 SNPs and at least one of the renal outcomes. We genotyped these SNPs in a second set of 472 white patients from 229 families with PKD1 and performed a joint analysis on both cohorts. Three SNPs continued to show suggestive/significant association with eGFR at the Dickkopf 3 (DKK3) gene locus; no SNPs significantly associated with renal survival. DKK3 antagonizes Wnt/β-catenin signaling, which may modulate renal cyst growth. Pending replication, our study suggests that genetic variation of DKK3 may modify severity of ADPKD resulting from PKD1 mutations.
Autosomal dominant polycystic kidney disease ( ADPKD) is the most common monogenic kidney disease worldwide, affecting one in 500 to 1000 births.1,2 It is characterized by focal development of renal cysts in an age-dependent manner. Typically, only a few renal cysts are clinically detectable during the first three decades of life; however, by the fifth decade, tens of thousands of renal cysts of different sizes can be found in most patients.3 Progressive cyst expansion with age leads to massive enlargement and distortion of the normal architecture of both kidneys and, ultimately, ESRD in most patients. ADPKD is also associated with an increased risk for cardiac valvular defects, colonic diverticulosis, hernias, and intracranial arterial aneurysms. Overall, ADPKD accounts for approximately 5% of ESRD in North America.2
Mutations of PKD1 and PKD2 respectively account for approximately 85% and approximately 15% of linkage-characterized European families. Polycystin-1 (PC-1) and PC-2, the proteins encoded by PKD1 and PKD2, respectively, function as a macromolecular complex and regulate multiple signaling pathways to maintain the normal tubular structure and function.1 Monoclonal expansion of individual epithelial cells that have undergone a somatic “second hit” mutation, resulting in biallelic inactivation of either PKD1 or PKD2, seems to provide a major mechanism for focal cyst initiation,4 possibly through the loss of polycystin-mediated mechanosensory function in the primary cilium.5 In addition, a large prospective, observational study indicated that renal cysts in ADPKD expand exponentially with increasing age, and patients with large polycystic kidneys are at higher risk for developing kidney failure6; however, the key factors that modulate renal disease progression in ADPKD remain incompletely understood.
Renal disease severity in ADPKD is highly variable, with the age of onset of ESRD ranging from childhood to old age.7–11 A strong genetic locus effect has been noted in ADPKD. Adjusted for age and gender, patients with PKD1 have larger kidneys and earlier onset at ESRD than patients with PKD2 (mean age at ESRD 53.4 versus 72.7 years, respectively).8,9 By contrast, a weak allelic effect (based on the 5′ versus 3′ location of the germline mutations) on renal disease severity may be present for PKD110 but not PKD2.11 Marked intrafamilial variability in renal disease is well documented in ADPKD and suggests a strong modifier effect.10–15 In an extreme example, large polycystic kidneys were present in utero in one of a pair of dizygotic twins affected with the same germline PKD1 mutation, whereas the kidneys of the co-twin remained normal at 5 years of age.12 Several studies have quantified the role of genetic background in the phenotypic expression of ADPKD. In a comparison of monozygotic twins and siblings, greater variance in the age of onset of ESRD in the siblings supported a role for genetic modifiers.13 Two other studies of intrafamilial disease variability in PKD1 have estimated that genetic factors may account for 32 to 42% of the variance of creatinine clearance before ESRD and 43 to 78% of the variance in age at ESRD.14,15 The magnitude of the modifier gene effect from these studies suggests that mapping such factors is feasible. Here, we report the results of an association study of modifier genes for PKD1 renal disease severity.
Results
Genotype and Phenotype Data
We designed a customized Illumina array to study 173 candidate genes with 1536 single-nucleotide polymorphisms (SNPs; Table 1; see the Concise Methods section and supplemental information), including 100 ancestry informative markers (AIM) for European ancestry.16,17 We selected our candidate genes on the basis of the known pathophysiology of renal disease progression in ADPKD, including genes involved in xenobiotic metabolism, DNA repair, BP control, and tissue fibrotic response. From our microarray gene expression study,18 we also selected genes from pathways that might modulate renal cyst growth. They include genes from pathways that regulate intracellular calcium and cAMP concentrations, Wnt/β-catenin, pleiotropic growth factor/receptor tyrosine kinase (e.g., IGF/IGF1R, EGF/EGFR, FGF/FGFR, PI3K/Akt/mTOR) and G-protein–coupled receptor (e.g., PTGER2) signaling, and angiogenesis. A total of 794 patients from 227 families with PKD1 (cohort 1) and 1495 SNPs that passed all quality control (QC) measures were analyzed (see the Study Cohorts section). Forty-five percent were male, 7.2% were singleton cases, and 38% had ESRD. There were no gender differences in their clinical characteristics (Supplemental Table S1). Overall, 22.7, 23.4, 12.2, and 41.7% of our patients were classified as having stages 1, 2, 3, and 4/5 chronic kidney disease (CKD), respectively (Supplemental Table S2). Comparing the distribution of patients with different CKD stages by study sites revealed heterogeneity of renal disease severity. Some centers (Newfoundland, Aachen, and Nicosia) contributed more patients with stage 1 CKD, whereas others (Brussels, Leiden, Barcelona, Oviedo, and Greece) contributed a very high proportion of patients with stage 5 CKD (Supplemental Table S3). Table 2 provides a summary of the clinical characteristics of our study patients by site.
Table 1.
Biological candidate genes studied (n = 173)
| Candidate Genes (Grouped by Function or Pathways) |
|---|
| Xenobiotic metabolism (n = 8) |
| ABCB1, AHR, CYP2D6, CYP3A4, GSTM1, GSTP1, NAT1, NAT2 |
| DNA repair (n = 19) |
| ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, ERCC6, OGG1, PGBD3, SOD1, SOD2, TP53, TP73, XPA, XRCC1, XRCC2, XRCC4, XRCC5, MTHFR, MTR |
| Aging (n = 3) |
| KL, SIRT1, WRN |
| Angiogenesis (n = 13) |
| ANGPT2, ANGPTL4, CTGF, EPHB4, ERAP1, FIGF, KDR, SERPINE1, HIF1A, VHL, VEGFA, VEGFB, VEGFC |
| Apoptosis (n = 5) |
| CASP8, TNFRSF1A, TNFRSF1B, TNFRSF21, TNFRSF25 |
| Intracellular calcium signaling (n = 9) |
| CALM2, NFAT5, NFATC1, NFATC4, PLCE1, PPP3CA, PPP3CB, PPP3CC, PPP3R1 |
| Cilia-associated genes (n = 14) |
| BBS10, BBS4, BBS7, GLI2, IQCB1, LGALS3, NEK1, NEK8, NPHP3, NPHP4, PDGFRA, PKD2, TRIM32, TTC8 |
| Canonical Wnt signaling (n = 14) |
| APC, DKK2, DKK3, FRZB, GSK3B, INVS, MYC, RSPO1, RSPO3, SFRP4, WNT11, WNT2, WNT5B, WNT9A |
| PI3K/Akt/mTOR signaling (n = 20) |
| AKT1, AKT2, AKT3, PTEN, PRKAB1, PRKAG1, FRAP1, IGF1, IGF1R, IRS1, IRS2, PIK3CA, PIK3R1, PRKAA1, RHEB, RPS6KA1, STK11, TSC1, TSC2, ULK2 |
| MAPK signaling (n = 19) |
| BRAF, DUSP1, DUSP8, EGF, EGFR, FGF18, FGFR1, FGFR3, FGFR4, IL1R1, MAP2K1, MAP3K3, MAP3K4, MAP3K6, MAPK1, PDGFB, PDGFRB, RAF1, TGFA |
| JAK-STAT signaling (n = 17) |
| IFNAR2, IL10RB, IL13, IL13RA1, IL13RA2, IL2RG, IL4, IL4R, IL6, SOCS3, SOCS5, STAT1, STAT2, STAT3, STAT5A, STAT5B, STAT6 |
| Renin-angiotensin system (n = 6) |
| REN, AGT, AGTR1, AGTR2, ACE, ACE2 |
| Prostaglandin signaling (n = 5) |
| ALOX12, PLA2G2A, PTGER2, PTGS1, PTGS2 |
| TGF-β signaling (n = 11) |
| BMP2, BMP7, DCN, GREM1, ID2, TGFB1, TGFB2, TGFBR1, TGFBR2, THBS1, THBS2 |
| Miscellaneous genes (n = 10) |
| ADCY3, CFTR, CSK, CXCL12, IL17D, IL33, IL8RA, ILK, PPARD, SPARC |
Some genes may be involved in multiple biological processes and pathways.
Table 2.
Patient characteristics (COHORT1) by study site
| Study Site | Patient | Birth Year | Age at Last Scr (years) | eGFR (ml/min) | % with ESRD | Age at ESRD (years) | Time to ESRD (years) | C1 | C2 | C3 | C4 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Toronto, Canada | 174 | 1952 ± 13 | 43.3 ± 11.4 | 51.7 ± 44.1 | 41.4 | 49.8 ± 8.3 | 43.3 ± 11.4 | 0.0050 ± 0.0200 | 0.0006 ± 0.0200 | −0.0100 ± 0.0300 | 0.0020 ± 0.0200 |
| Newfoundland, Canada | 79 | 1957 ± 12 | 41.5 ± 11.5 | 70.7 ± 42.3 | 19.0 | 47.9 ± 8.8 | 41.5 ± 11.5 | 0.0200 ± 0.0100 | −0.0040 ± 0.0400 | 0.0040 ± 0.0300 | −0.0030 ± 0.0300 |
| Denver, CO | 216 | 1955 ± 16 | 40.0 ± 15.0 | 64.9 ± 45.0 | 31.5 | 51.4 ± 11.3 | 40.0 ± 15.0 | −0.0020 ± 0.0200 | 0.0030 ± 0.0200 | −0.0020 ± 0.0200 | −0.0030 ± 0.0200 |
| Brussels, Belgium | 21 | 1946 ± 19 | 46.7 ± 10.3 | 23.6 ± 35.5 | 71.4 | 48.6 ± 10.4 | 46.7 ± 10.3 | −0.0080 ± 0.0300 | 0.0060 ± 0.0100 | 0.0040 ± 0.0200 | −0.0030 ± 0.0300 |
| Sofia, Bulgaria | 22 | 1949 ± 13 | 45.6 ± 11.9 | 45.8 ± 31.1 | 40.0 | 54.4 ± 5.1 | 45.6 ± 11.9 | −0.0300 ± 0.0200 | 0.0100 ± 0.0200 | 0.0100 ± 0.0200 | −0.0060 ± 0.0200 |
| Leiden, Netherlands | 41 | 1945 ± 9.5 | 49.5 ± 8.7 | 27.6 ± 28.8 | 68.3 | 52.5 ± 6.5 | 49.5 ± 8.7 | 0.0060 ± 0.0200 | 0.0006 ± 0.0200 | −0.0100 ± 0.0100 | 0.0100 ± 0.0200 |
| Aachen, Germany | 12 | 1962 ± 14 | 39.3 ± 10.7 | 69.4 ± 48.6 | 33.3 | 50.5 ± 8.7 | 39.3 ± 10.7 | −0.0020 ± 0.0200 | 0.0010 ± 0.0100 | −0.0060 ± 0.0100 | 0.0050 ± 0.0200 |
| Modena, Italy | 28 | 1956 ± 13 | 43.8 ± 11.4 | 54.4 ± 44.0 | 28.6 | 49.4 ± 10.8 | 43.8 ± 11.4 | −0.0300 ± 0.0080 | −0.0050 ± 0.0200 | −0.0060 ± 0.0100 | −0.0090 ± 0.0200 |
| Barcelona, Spain | 21 | 1948 ± 9 | 46.0 ± 5.6 | 27.0 ± 35.0 | 76.2 | 46.6 ± 6.1 | 46.0 ± 5.6 | −0.0300 ± 0.0200 | −0.0020 ± 0.0200 | −0.0100 ± 0.0200 | 0.0080 ± 0.0100 |
| Oviedo, Spain | 22 | 1948 ± 8 | 54.4 ± 6.8 | 19.1 ± 20.4 | 82.0 | 53.7 ± 6.6 | 54.4 ± 6.8 | −0.0300 ± 0.0200 | −0.0090 ± 0.0100 | −0.0070 ± 0.0100 | 0.0060 ± 0.0200 |
| Kuopio, Finland | 119 | 1952 ± 16 | 44.5 ± 15.3 | 66.1 ± 37.4 | 21.0 | 54.3 ± 13.1 | 44.5 ± 15.3 | 0.0400 ± 0.0200 | 0.0200 ± 0.0200 | 0.0200 ± 0.0200 | 0.0010 ± 0.0200 |
| Nicosia, Cyprus | 20 | 1962 ± 14 | 40.6 ± 12.8 | 55.9 ± 44.0 | 40.0 | 50.4 ± 5.6 | 40.6 ± 12.8 | −0.0700 ± 0.0100 | 0.0020 ± 0.0100 | 0.000007 ± 0.0200 | −0.0100 ± 0.0200 |
| Athens, Greece | 19 | 1947 ± 9 | 50.5 ± 10.2 | 10.0 | 100.0 | 50.5 ± 10.2 | 50.5 ± 10.2 | −0.0400 ± 0.0200 | −0.0060 ± 0.0100 | 0.0040 ± 0.0100 | −0.0030 ± 0.0100 |
| All centers | 794 | 1953 ± 15 | 43.3 ± 13.2 | 55.1 ± 43.5 | 38.3 | 50.8 ± 9.5 | 43.3 ± 13.2 | 0.0020 ± 0.0300 | −0.0002 ± 0.0300 | −0.0010 ± 0.0200 | −0.0005 ± 0.0200 |
All continuous variables are mean ± SD. Scr, serum creatinine.
Analysis of Population Structure
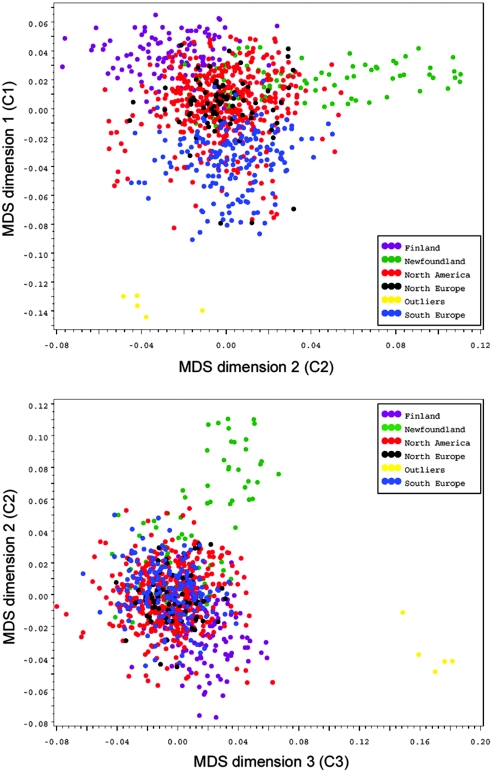
We used 100 AIMs for European ancestry16,17 and 308 tagSNPs (i.e., SNPs that were not in linkage disequilibrium (LD) with all other markers; r2 < 0.06) to test and control for population structure by the multidimensional scaling (MDS) method from PLINK 1.05.19 Population structure was captured by MDS dimensions 1 through 4 (C1 through C4) corresponding to different geographic sites (Table 2). Figure 1 shows the clustering of patients from different geographic sites by MDS dimensions. C1 separates patients along a northwest-southeast axis of European ancestry. North Americans represent an admixed group along this axis. C2 separates the Fins from Newfoundlanders. C3 and C4 provide the best separation and identification of five outliners who were subsequently shown to be black (Supplemental Figure S1). Differences of population structure as captured by C1 through C4 were evident by different CKD stages, reflecting patient admixture from different geographic sites (Supplemental Table S3). This heterogeneity is reflected in the patient characteristics by geographic site (Table 2).
Figure 1.
Detection and adjustment of population structure by multidimensional scaling (MDS). C1 separates patients along a northwest-southeast axis of their European ancestry. North Americans form an admixed group along this axis. C2 separates the Fins from the Newfoundlanders. Five outliers were subsequently identified to be black.
Genetic Associations with Renal Outcomes
Using the modeling framework detailed in the Statistical Analysis section, we found 12 SNPs with suggestive association (P = ∼0.005) with at least one of the two outcomes (Table 3). In general, the genotype QC of these SNPs was excellent, with marker missing rate ≤1%. We found the strongest associations from rs3750940 and rs12575803, both located in DKK3, with an identical P = 0.00019 for eGFR. Several SNPs at RHEB, PPP3R1, CALM2, PTGS2, IL1R1, and ERCC3 were weakly associated with either of the renal outcomes (P = ∼0.005). Three SNPs at both DKK3 and RHEB are in LD with r2 of 0.56 to 0.88 and 0.73 to 0.81, respectively. Two SNPs at CALM2, rs1693869 and rs815802, are also in moderate LD (r2 = 0.57). To evaluate these suggestive associations further, we genotyped them in a second cohort of 472 white patients with PKD1 from Oxford, England, and Rochester, Minnesota (cohort 2; see the Study Cohorts section). There were more female patients from both sites, and the Oxford cohort was more enriched with patients with ESRD (Supplemental Table S4). We performed similar analysis as before except that only family relationship was adjusted for eGFR, and family relationship was adjusted for renal survival. We found that only two SNPs at DKK3 (rs3750940 and rs7104941) continued to show suggestive associations (P = ∼0.05) for eGFR (Table 4). We then combined the two patient cohorts (n = 1266) for a joint analysis and found P = 8.0 × 10−5 for rs3750940 and P = ∼5 × 10−4 for rs7104941 and rs12575803 all at DKK3 (Table 5). From the EFFECT estimates of the generalized estimating equations (GEE) model, we found that each copy of the risk allele from the three associated DDK3 SNPs is associated with a difference of eGFR of approximately 7 to 8 ml/min. We also analyzed the aforementioned SNPs in the combined patient cohort using Merlin, which uses a variance components association method and adjusts for family relationship of related individuals using kinship coefficients. Consistent with the results by GEE, we found the same three SNPs from DKK3 continued to show suggestive/significant association with eGFR (Table 6). The SNP rs3750940 provides the strongest association at P = 4.6 × 10−5 and accounts for 1.4% of the total variance of eGFR.
Table 3.
SNPs with suggestive associations (COHORT1, n = 794)
| SNP ID | Gene | Chromosome | Physical Location (bp) | eGFR Pa | Renal Survival Pb | Alleles | Reference Allele | RAFc | HapMap RAFd | HWE P | Marker Missing (%) | GenCall Scoree |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3750940 | DKK3 | 11 | 11979474 | 0.00019 | 0.22 | A/G | G | 0.20 | 0.21 | 0.56 | 0.24 | 0.82 |
| rs7104941 | DKK3 | 11 | 11977192 | 0.0027 | 0.27 | G/A | A | 0.19 | 0.17 | 0.24 | 0.24 | 0.74 |
| rs12575803 | DKK3 | 11 | 11981152 | 0.00019 | 0.55 | C/A | A | 0.14 | 0.14 | 0.59 | 0.48 | 0.92 |
| rs875588 | RHEB | 7 | 150799763 | 0.0017 | 0.27 | G/A | A | 0.48 | 0.48 | 0.57 | 0.12 | 0.83 |
| rs3753151 | RHEB | 7 | 150815918 | 0.0051 | 0.30 | G/A | A | 0.53 | 0.57 | 0.37 | 0.72 | 0.78 |
| rs6972955 | RHEB | 7 | 150802595 | 0.0022 | 0.47 | C/A | A | 0.44 | 0.48 | 0.95 | 0.24 | 0.81 |
| rs6546365 | PPP3R1 | 2 | 68328790 | 0.0015 | 0.012 | G/A | A | 0.48 | 0.43 | 0.67 | 1.10 | 0.77 |
| rs1693869 | CALM2 | 2 | 47255568 | 0.23 | 0.0034 | G/C | C | 0.12 | 0.13 | 0.80 | 0.00 | 0.86 |
| rs815802 | CALM2 | 2 | 47245553 | 0.72 | 0.0046 | A/G | G | 0.09 | 0.08 | 0.43 | 0.24 | 0.86 |
| rs2206593 | PTGS2 | 1 | 184909052 | 0.53 | 0.0016 | G/A | A | 0.08 | 0.06 | 0.69 | 0.60 | 0.85 |
| rs3917225 | IL1R1 | 2 | 102135734 | 0.21 | 0.0019 | A/G | G | 0.48 | 0.46 | 0.83 | 0.24 | 0.86 |
| rs4150471 | ERCC3 | 2 | 127751009 | 0.52 | 0.0036 | G/A | A | 0.28 | 0.23 | 1.00 | 0.36 | 0.90 |
HWE, Hardy-Weinberg equilibrium; RAF, reference allele frequency.
aAdjusted for age, MDS dimensions C1 through C4, and family relationship by GEE.
bCox proportional hazards model was used for renal survival (absence of ESRD) analysis; adjusted for MDS dimensions C1 through C4 and family relationship.
cRAF was derived from a random draw of one genotyped individual from each family.
dRAF from the HapMap database for European population.
eThe GenCall scores ≥0.7 indicate well-behaving genotypes.
Table 4.
Replication of SNPs with suggestive associations (COHORT2, n = 472)
| SNP ID | Gene | Chromosome | Physical Location (bp) | eGFR Pa | Renal Survival Pb | Alleles | Reference Allele | RAFc | HapMap RAFd | HWE P | Marker Missing (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3750940 | DKK3 | 11 | 11979474 | 0.067 | 0.43 | A/G | G | 0.17 | 0.21 | 0.05 | 0.0 |
| rs7104941 | DKK3 | 11 | 11977192 | 0.070 | 0.23 | G/A | A | 0.16 | 0.17 | 0.11 | 0.0 |
| rs12575803 | DKK3 | 11 | 11981152 | 0.22 | 0.70 | C/A | A | 0.12 | 0.14 | 1.00 | 4.3 |
| rs875588 | RHEB | 7 | 150799763 | 0.25 | 0.97 | G/A | A | 0.52 | 0.48 | 0.13 | 1.7 |
| rs3753151 | RHEB | 7 | 150815918 | 0.39 | 0.74 | G/A | A | 0.58 | 0.57 | 0.19 | 0.0 |
| rs6972955 | RHEB | 7 | 150802595 | 0.11 | 0.61 | C/A | A | 0.42 | 0.42 | 0.06 | 0.0 |
| rs6546365 | PPP3R1 | 2 | 68328790 | 0.36 | 0.22 | G/A | A | 0.44 | 0.43 | 0.59 | 0.0 |
| rs1693869 | CALM2 | 2 | 47255568 | 0.59 | 0.84 | G/C | C | 0.12 | 0.13 | 0.38 | 0.0 |
| rs815802 | CALM2 | 2 | 47245553 | 0.26 | 0.47 | A/G | G | 0.10 | 0.08 | 0.38 | 0.0 |
| rs2206593 | PTGS2 | 1 | 184909052 | 0.98 | 0.84 | G/A | A | 0.05 | 0.06 | 0.30 | 0.0 |
| rs3917225 | IL1R1 | 2 | 102135734 | 0.69 | 0.76 | A/G | G | 0.48 | 0.46 | 0.95 | 0.0 |
| rs4150471 | ERCC3 | 2 | 127751009 | 0.059 | 0.96 | G/A | A | 0.27 | 0.23 | 0.51 | 0.4 |
HWE, Hardy-Weinberg equilibrium; RAF, reference allele frequency.
aAdjusted for age and family relationship by GEE.
bCox proportional hazards model for renal survival analysis; adjusted for family relationship.
cRAF was derived from a random draw of one genotyped individual from each family.
dRAF from the HapMap database for European population.
Table 5.
Combined analysis of cohorts 1 and 2 (n = 1266) by GEE
| SNP ID | Gene | Chromosome | Physical Location (bp) | Reference Allele | RAFa | eGFR Pb | Effect (95% CI)c | Renal Survival Pd | Effect (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| rs3750940 | DKK3 | 11 | 11979474 | G | 0.21 | 8.0 × 10−5 | −8.000 (−12.000 to −4.000) | 0.095 | 0.090 (−0.020 to 0.200) |
| rs7104941 | DKK3 | 11 | 11977192 | A | 0.20 | 6.2 × 10−4 | −7.300 (−12.000 to −3.100) | 0.059 | 0.110 (−0.004 to 0.220) |
| rs12575803 | DKK3 | 11 | 11981152 | A | 0.15 | 4.2 × 10−4 | −8.700 (−13.000 to −3.900) | 0.70 | 0.025 (−0.100 to 0.150) |
| rs875588 | RHEB | 7 | 150799763 | A | 0.50 | 0.046 | 3.600 (0.060 to 7.000) | 0.56 | −0.025 (−0.110 to 0.057) |
| rs3753151 | RHEB | 7 | 150815918 | A | 0.56 | 0.073 | 3.300 (−0.310 to 7.000) | 0.79 | −0.011 (−0.095 to 0.072) |
| rs6972955 | RHEB | 7 | 150802595 | A | 0.44 | 0.050 | 3.600 (0.003 to 7.200) | 0.48 | −0.028 (−0.100 to 0.049) |
| rs6546365 | PPP3R1 | 2 | 68328790 | A | 0.40 | 2.9 × 10−3 | 4.700 (1.600 to 7.800) | 5.7 × 10−3 | −0.100 (−0.170 to −0.030) |
| rs1693869 | CALM2 | 2 | 47255568 | C | 0.14 | 0.26 | −2.600 (−7.300 to 2.000) | 0.014 | 0.160 (0.030 to 0.280) |
| rs815802 | CALM2 | 2 | 47245553 | G | 0.11 | 0.59 | −1.400 (−6.700 to 3.800) | 8.6 × 10−3 | 0.180 (0.040 to 0.310) |
| rs2206593 | PTGS2 | 1 | 184909052 | A | 0.05 | 0.37 | 3.000 (−3.600 to 9.700) | 0.075 | −0.140 (−0.290 to 0.014) |
| rs3917225 | IL1R1 | 2 | 102135734 | G | 0.47 | 0.20 | −2.300 (−5.700 to 1.200) | 0.013 | 0.100 (0.020 to 0.180) |
| rs4150471 | ERCC3 | 2 | 127751009 | A | 0.27 | 0.73 | −0.730 (−4.800 to 3.400) | 0.033 | 0.100 (0.008 to 0.200) |
CI, confidence interval; RAF, reference allele frequency.
aRAF was derived from a random draw of one genotyped individual from each family.
bAdjusted for age and family relationship by GEE.
cProvides an estimate of the effect size of the reference allele under an additive model.
dCox proportional hazards model for renal survival analysis, adjusted for family relationship.
Table 6.
Combined analysis of cohorts 1 and 2 (n = 1266) by Merlin
| SNP ID | Gene | Chromosome | Physical Location (bp) | Reference Allele | RAFa | eGFR Pb | H2 (%)c | Renal Survival Pd | H2 (%) |
|---|---|---|---|---|---|---|---|---|---|
| rs3750940 | DKK3 | 11 | 11979474 | G | 0.21 | 4.6 × 10−5 | 1.40 | 0.24 | 0.13 |
| rs7104941 | DKK3 | 11 | 11977192 | A | 0.20 | 1.4 × 10−4 | 1.30 | 0.12 | 0.23 |
| rs12575803 | DKK3 | 11 | 11981152 | A | 0.15 | 6.7 × 10−4 | 0.95 | 0.92 | 0.00 |
| rs875588 | RHEB | 7 | 150799763 | A | 0.50 | 0.14 | 0.21 | 0.97 | 0.00 |
| rs3753151 | RHEB | 7 | 150815918 | A | 0.56 | 0.20 | 0.16 | 0.91 | 0.00 |
| rs6972955 | RHEB | 7 | 150802595 | A | 0.44 | 0.052 | 0.37 | 0.77 | 0.01 |
| rs6546365 | PPP3R1 | 2 | 68328790 | A | 0.40 | 0.0081 | 0.57 | 0.052 | 0.35 |
| rs1693869 | CALM2 | 2 | 47255568 | C | 0.14 | 0.12 | 0.27 | 0.044 | 0.47 |
| rs815802 | CALM2 | 2 | 47245553 | G | 0.11 | 0.25 | 0.15 | 0.020 | 0.63 |
| rs2206593 | PTGS2 | 1 | 184909052 | A | 0.05 | 0.44 | 0.06 | 0.19 | 0.17 |
| rs3917225 | IL1R1 | 2 | 102135734 | G | 0.47 | 0.11 | 0.23 | 0.047 | 0.38 |
| rs4150471 | ERCC3 | 2 | 127751009 | A | 0.27 | 0.63 | 0.02 | 0.037 | 0.95 |
RAF, reference allele frequency.
aRAF was derived from a random draw of one genotyped individual from each family.
bRank transformed and adjusted for age and family relationship by the variance components association method in Merlin.
cProportion of the variance estimated to be accounted for by the marker.
dCox proportional hazards model for renal survival analysis, rank transformed and adjusted for family relationship by the variance components association method.
DKK3 Expression in PKD1 Renal Cysts
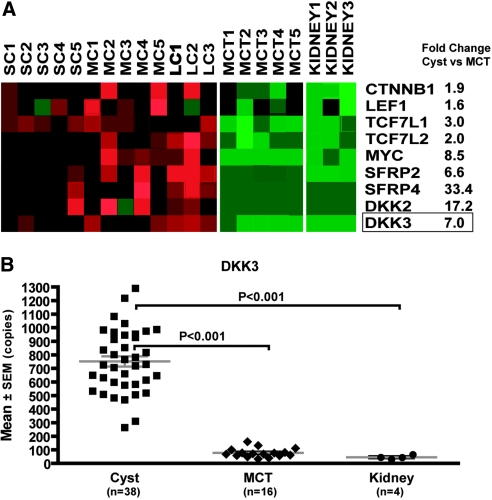
Using microarray global gene profiling, we previously documented aberrant activation of the Wnt signaling pathway in human PKD1 renal cysts.18,20 Using the same database, we examined the gene expression profile of DKK3. Consistent with aberrant Wnt activation, we found a number of target genes (β-catenin [CTNNB1], LEF1, TCF7, and MYC) and negative modulators including DKK3 and the secreted frizzled-related proteins showed higher expression in PKD1 renal cysts compared with minimally cystic control tissue from the same kidneys (Figure 2A). Using real-time reverse transcriptase–PCR (RT-PCR), we further validated the differential expression pattern of DKK3 in an expanded number of renal cysts and control tissue samples (Figure 2B).
Figure 2.
Differential expression of DKK3 in human PKD1 renal cysts. (A) Differential expression of selected component and target genes suggesting aberrant activation of Wnt/β-catenin signaling pathway in renal cysts (n = 13) compared with minimally cystic tissues (MCT; n = 5) from human PKD1 kidneys (false-discovery rate <0.5%). The expression pattern of these genes in MCT and normal renal cortical tissue (Kidney; n = 3) is very similar. Upregulated genes are shown in red, and downregulated genes are shown in green. (B) Real-time RT-PCR analysis of DKK3 in an expanded sample set (Cyst = 38; MCT = 16; Kidney = 4). Data are means ± SEM; one-way ANOVA with Tukey multiple comparisons posttest. SC, small cysts; MC, medium cysts; LC, large cysts.
Discussion
We present here the largest study of PKD1 genetic modifiers of renal disease severity reported to date. Using high-throughput SNP genotyping, our study provided a comprehensive scan for genetic association of common variation in 173 biological candidate genes. After adjusting for population structure and performing statistical analysis that accommodated for both family- and population-based data, we found 12 SNPs with suggestive associations in cohort 1; however, only three SNPs continued to show suggestive/significant associations in the joint analysis with an expanded sample size. These SNPs, located at DKK3, are in moderate to high LD with each other. Our results were highly consistent using two different methods of quantitative trait analysis that accommodate family- and population-based association. The SNP rs3750940 provided the strongest association with a P value that approaches (8.0 × 10−5 by GEE) or exceeds (4.6 × 10−5 by Merlin) the threshold of statistical significance that corrects for multiple testing (4.7 × 10−5; see the Statistical Analysis section). Given that these associated SNPs reside in intronic regions, it is unclear whether any of them may be functional. Rather, it is likely that they are in LD with the causal variant(s) elsewhere, which may alter the expression or function of DKK3. We found that each copy of the risk allele from rs3750940 was associated with a difference in eGFR of approximately 7 to 8 ml/min and that this marker accounts for 1.4% of the total variance of eGFR. Pending replication, our study suggests that genetic variation of DKK3 may modify renal disease progression in patients with PKD1.
WNTs comprise an evolutionarily conserved family of growth factors that are critically involved in kidney development and regeneration, by binding to Frizzled and LRP5/6 receptors to stabilize β-catenin to initiate T cell factor/lymphocyte enhancer factor (TCF/LEF)-dependent gene transcription. Dysregulation of WNT signaling contributes to a number of human diseases, including PKD.21 Specifically, aberrant activation of β-catenin in transgenic mice has been shown to cause PKD.22,23 Moreover, nuclear translocation of the C-terminal tail of PC-1 has been shown to inhibit β-catenin/TCF-dependent gene transcription in vitro, and the loss of the PC-1 C-terminal tail from truncating PKD1 mutations may contribute to the aberrant Wnt activation in ADPKD.20 DKKs are a family of secreted glycoproteins that function as antagonists to downregulate WNT signaling.24 Consistent with their function, we found upregulation of DKK2 and DKK3 expression in human PKD1 renal cysts. Our findings, if confirmed, suggest that therapeutic antagonism of WNT signaling may be useful in ADPKD.
Despite testing a large number of candidate genes, we found only three SNPs in DKK3 that showed suggestive/significant association for eGFR but not renal survival. We interpret these findings to suggest one or more of the following: (1) Different genetic factors may modify each of the two renal outcomes; (2) renal survival from ESRD may not be as sensitive a measure of disease severity as eGFR; (3) some candidate SNPs we tested might indeed modify PKD1 renal disease modestly, but our sample size was not powered to detect such effects; and (4) most SNPs from our candidate genes were not associated with the renal outcomes. Future studies with larger patient samples may help to clarify these issues. At the same time, these findings suggest that the candidate gene approach may not be the best means to identify genetic modifiers given our current gap of knowledge on the mechanisms of renal disease progression in ADPKD and that an agnostic approach such as that taken by genome-wide association study (GWAS) is strongly justified (see the next paragraph).
The search for susceptibility/modifier genes for complex traits has until recently been fraught with problems.25 Although many putative loci were identified by candidate gene studies, few were reproducible in subsequent studies. A recent literature review showed that only six of 166 reported associations were replicated in follow-up studies.26 Many of these associations are likely spurious, because they were identified from studies of small sample size without adjustment for population stratification and multiple testing. Other reasons for the lack of reproducibility include genetic and etiologic heterogeneity, variable LD between the tested and causative variant, and false-negative results from inadequately powered studies. Minimizing phenotypic heterogeneity, robust QC for genotyping, genomic measures for population stratification, and the use of large sample sets for initial detection and follow-up replication all are important parameters for successful association studies.25,26 The success of GWAS has been demonstrated in many common medical conditions, including bipolar disorder, macular degeneration, long QT syndrome, coronary artery disease, diabetes, rheumatoid arthritis, and multiple forms of cancer, among others.27–34 To date, more than 150 risk loci have been identified for more than 60 common diseases and traits.35 The results from these studies have revealed new insights in disease pathobiology and possible therapeutic approaches. In general, most of the robust risk loci identified are associated with heterozygote odds ratios of 1.2 to 2.2.35 For a complex trait such as PKD1 renal disease variability, a sample size of 3000 to 4000 patients may be required to provide a properly powered GWAS to detect multiple loci with similar effect size. Future collaboration through an international network of research centers is essential for the realization of this promising approach.
Concise Methods
Study Cohorts
The study cohort for our candidate gene array study (cohort 1) comprised 890 patients from linkage-characterized families with PKD1 and 50 singleton patients with known PKD1 mutations from 13 sites. Clinical review excluded 19 affected children who were younger than 14 years, two unaffected individuals, and four patients from a family of mixed ethnicity (n = 4). After the completion of analysis for cohort 1, we genotyped the 12 most associated SNPs that passed all QC measures identified from cohort 1 in a second cohort of 472 patients with PKD1 provided by Dr. Peter Harris (cohort 2). They were self-reported white individuals recruited from 233 families (with 26% singleton cases) from Oxford, England, and Rochester, Minnesota, and were characterized to have PKD1 by DNA linkage or mutations. The within-family relationship of all of the study patients are detailed in Supplemental Table S5. The institutional human subject review board or ethics committee from each study site approved the research protocol used in this study.
Clinical Assessment and Study Outcomes
All study patients were confirmed to have PKD1 by DNA linkage, documentation of pathogenic mutations, or age-dependent ultrasound criteria.36 We reviewed their demographic information including age, gender, ethnicity, study center, and family relationships to other affected relatives recruited in our study. Patients with a concomitant renal disease (e.g., diabetes, glomerulonephritis) or at the extreme of body weights (outside ±2 SD of the population mean) were excluded. For patients without ESRD, their last available serum creatinine level was used to calculate eGFR. For patients with ESRD, a default value of 10 ml/min was assigned as their eGFR, and their age at ESRD was used for renal survival analysis. We analyzed two primary renal outcomes: (1) eGFR as measured by the abbreviated Modification of Diet in Renal Disease (MDRD) equation37 and (2) renal survival from ESRD.
SNP Genotyping
We used a customized Illumina array and the GoldenGate assay to genotype cohort 1. All of the SNPs used in this study underwent bioinformatics evaluation to score their genotyping suitability. Most were tagSNPs (i.e., pair-wise r2 >0.8) selected from the HapMap phase II CEU data, but approximately 5% of them were nonsynonymous coding SNPs identified from the SeattleSNP and dbSNP databases. We also genotyped 12 most associated SNPs identified from cohort 1 in a second independent set of patients with PKD1 (cohort 2) using the MassARRAY iPlex assay (Sequenom). All SNPs underwent bioinformatic evaluation to design and score the PCR primers for multiplex genotyping. On the basis of 50 random samples genotyped by both assays, the concordant rate between the Illumina and Sequenom platforms was 99.3%. All DNA samples used were quantified by the picogreen method.
QC of SNP Genotyping
There was no genotype for 18 (1.2%) SNPs for technical reasons. In addition, an SNP was excluded when >30% of the genotypes were missing (n = 9). Minor allele frequency was calculated using one randomly selected individual from each family (n = 227). We tested for departure from Hardy-Weinberg equilibrium, because genotype problems are the most common cause for deviation from Hardy-Weinberg equilibrium. The minimum minor allele frequency was 0.01 (removed n = 14). A total of 1495 SNPs were used in the analysis. The mean concordance rate for 20 blind duplicates was 99.4%.
Fifteen patients were excluded for the following reasons: Inconsistency between reported and genotyped gender (n = 1), marker Mendelian errors >5% (n = 8), and average heterozygosity >0.4 (potentially indicative of sample contamination; n = 6). Using PLINK, we estimated pair-wise identify-by-descent sharing to determine the pair-wise relationships of our patients.19 We identified and excluded 10 samples with identical identify-by-descent (three patients from three monozygotic twin pairs and one family [n = 7] studied by two different sites). We also excluded a family of five identified as outliers by our population structure analysis (Figure 1) that was subsequently confirmed to be black. Finally, 91 samples (eight from Denver; three from Spain; one from the Netherlands; 14 from Cyprus; 12 from Greece; and 53 from Bulgaria) with >10% missing SNP genotypes were excluded. Most of the latter DNA samples were old or of low concentration. According to the technical notes by Illumina, GenCall scores <0.2 indicate poor quality SNPs and scores ≥0.7 “usually report well-behaving genotypes.” In this study, we excluded all SNPs with GenCall scores <0.3.
Population Structure
The population structure was tested by the MDS from PLINK 1.05 using 408 tagSNPs whose maximum pair-wise LD (measured by r2) with all other SNPs in the customized array was <0.06. One hundred of these tagSNPs were specifically selected from the AIM panel by Price et al.,16 which was derived from several large GWAS databases of North American white populations of well-defined European ancestry. Eighty-five of these AIMs were highly specific for differentiation between northwest and southeast European ancestry and 15 AIMs for differentiation between southeast European and Ashkenazi Jewish ancestry (see Figure 1 and Supplemental Figure S1). The dimensions derived from MDS were used as covariates in the GEE model for eGFR and time to ESRD.
Microarray Analysis and Real-Time RT-PCR
We previously performed microarray global gene profiling of renal cysts compared with minimally cystic control tissue from human PKD1 kidneys.18,20 Using the same database, we examined the gene expression profile of DKK3 and other selected genes from the WNT signaling pathway in PKD1 renal cysts compared with minimally cystic control tissues from the same kidneys. To validate that DKK3 is differentially expressed in PKD1 renal cysts, we performed real-time RT-PCR in expanded PKD1 renal cysts and control samples using established methods.18
Statistical Analysis
We analyzed two primary renal outcomes: (1) eGFR and (2) renal survival from ESRD. Because patients with ESRD were given a default eGFR of 10 ml/min, eGFR was bimodally distributed. To perform appropriate analysis of this trait, we first fit the data using the Tobit model,38 a regression model for truncated or censored dependent variables. Next, the residuals from the Tobit model were used in a GEE model,39 to account for the relatedness among individuals within the same family. For renal survival, we used the variable “time to ESRD” (defined by age at ESRD for patients with ESRD and age at last serum creatinine measurement for patients without ESRD). We fitted a Cox proportional hazards model for renal survival analysis (Supplemental Figure S2).40 The deviance residuals from this model were used in another GEE model to account for the relatedness among individuals. For both outcomes, we also used the first four dimensions from MDS (C1 through C4) to correct for population structure. An additive genetic model was used by coding the genotypes to 0, 1, and 2 to represent the number of minor alleles. All of these analyses were performed using SAS 9.1.3 (SAS Institute, Cary, NC). We also performed the same analysis as in GEE using Merlin 1.1.2, which uses a variance components association method to adjust for the family relationship of related individuals using kinship coefficients.41,42 To deal with multiple testing in our study, we applied the program SNPSpD.43 SNPSpD takes into account the LD relationships of the SNPs to provide an effective number of independent markers (i.e., markers not in LD with each other) and the significant threshold after correcting for multiple comparisons of these independent markers.43,44 Of the 1495 SNPs used for cohort 1, the effective number of independent markers was 1071. To keep the type I error rate at 5%, the significant p value threshold should be 0.05/1071, or approximately 4.7 × 10−5.
Disclosures
None.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Canadian Institute of Health Research (CIHR; MOP77806) to Y.P.; a Canada Research Chair in Genetics of Complex Diseases to A.D.P.; and the CIHR Distinguished Scientist Award to P.P.
We thank Kairong Wang, Elizabeth Dicks, and Christina Heyer for superb assistance and the Centre for Applied Genomics (Toronto, Ontario, Canada) for the genotyping and all of the participants of the study.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1. Harris P, Torres V: Polycystic kidney disease. Annu Rev Med 60: 321–337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. US Renal Data System: Excerpts from the USRDS 2004 Annual Data Report. Am J Kidney Dis 45: S1–S280, 2005 [Google Scholar]
- 3. Grantham J, Geiser J, Evan A: Cyst formation and growth in autosomal dominant polycystic kidney disease. Kidney Int 31: 1145–1152, 1987 [DOI] [PubMed] [Google Scholar]
- 4. Pei Y: A “two-hit” model of cystogenesis in autosomal dominant polycystic kidney disease? Trends Mol Med 7: 151–156, 2001 [DOI] [PubMed] [Google Scholar]
- 5. Nauli S, Rossetti S, Kolb R, Alenghat F, Consugar M, Harris P, Ingber D, Loghman-Adham M, Zhou J: Loss of polycystin-1 in human cyst-lining epithelia leads to ciliary dysfunction. J Am Soc Nephrol 17: 1015–1025, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Grantham J, Torres V, Chapman A, Guay-Woodford L, Bae KT, King B, Wetzel L, Baumgarten D, Kenney P, Harris P, Klahr S, Bennett W, Hirschman G, Meyers C, Zhang XL, Zhu F, Miller J: Volume progression in polycystic kidney disease. N Engl J Med 354: 2122–2130, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Parfrey PS, Bear JC, Morgan J, Cramer BC, McManamon PJ, Gault MH, Churchill DN, Singh M, Hewitt R, Somlo S, Reeders S: The diagnosis and prognosis of autosomal dominant polycystic kidney disease. N Engl J Med 323: 1085–1090, 1990 [DOI] [PubMed] [Google Scholar]
- 8. Hateboer N, v Dijk MA, Bogdanova N, Coto E, Saggar-Malik AK, San Millan JL, Torra R, Breuning M, Ravine D: Comparison of phenotypes of polycystic kidney disease types 1 and 2. European PKD1-PKD2 Study Group. Lancet 353: 103–107, 1999 [DOI] [PubMed] [Google Scholar]
- 9. Harris PC, Bae KT, Rossetti S, Torres VE, Grantham JJ, Chapman AB, Guay-Woodford LM, King BF, Wetzel LH, Baumgarten DA, Kenney PJ, Consugar M, Klahr S, Bennett WM, Meyers CM, Zhang Q, Thompson PA, Zhu F, Miller JP, CRISP Consortium : Cyst number but not the rate of cystic growth is associated with the mutated gene in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 17: 3013–3019, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Rossetti S, Burton S, Strmecki L, Pond G, San Millan J, Zerres K, Barratt TM, Ozen S, Torres V, Bergstralh EJ, Winearls C, Harris PC: The position of the polycystic kidney disease 1 gene mutation correlates with the severity of renal disease. J Am Soc Nephrol 13: 1230–1237, 2002 [DOI] [PubMed] [Google Scholar]
- 11. Magistroni R, He N, Wang KR, Andrew R, Johnson A, Gabow P, Dicks E, Parfrey P, Torra R, San-Millan J, Coto E, v Dijk M, Breuning M, Peters D, Bogdanova N, Ligabue G, Albertazzi A, Hateboer N, Demetriou K, Pierides A, Deltas C, St. George-Hyslop P, Ravine D, Pei Y: Genotype-renal function correlation in type 2 autosomal dominant polycystic kidney disease. J Am Soc Nephrol 14: 1164–1174, 2003 [DOI] [PubMed] [Google Scholar]
- 12. Peral B, Ong A, San Millan J, Gamble V, Rees L, Harris PC: A stable, nonsense mutation associated with a case of infantile onset polycystic kidney disease 1. Hum Mol Genet 5: 539–542, 1996 [DOI] [PubMed] [Google Scholar]
- 13. Persu A, Duyme M, Pirson Y, Lens X, Messiaen T, Breuning M, Chauveau D, Levy M, Grunfeld J, Devuyst O: Comparison between siblings and twins supports a role for modifier genes in ADPKD. Kidney Int 66: 2132–2136, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Paterson AD, Magistroni R, He N, Wang K, Johnson A, Fain PR, Dicks E, Parfrey P, St. George-Hyslop P, Pei Y: Progressive loss of renal function is an age-dependent heritable trait in type 1 autosomal dominant polycystic kidney disease. J Am Soc Nephrol 16: 755–762, 2004 [DOI] [PubMed] [Google Scholar]
- 15. Fain P, McFann K, Taylor M, Tison M, Johnson A, Reed B, Schrier R: Modifier genes play a significant role in the phenotypic expression of PKD1. Kidney Int 67: 1256–1267, 2005 [DOI] [PubMed] [Google Scholar]
- 16. Price A, Butler J, Patterson N, Capelli C, Pascali V, Scarnicci F, Ruiz-Linares A, Groop L, Saetta A, Korkolopoulou P, Seligsohn U, Waiszewska A, Schirmer C, Ardlie K, Ramos A, Nemesh J, Arbeitman L, Goldstein D, Reich D, Hirschhorn J: Discerning the ancestry of European Americans in genetic association studies. PLoS Genet 4: 9–17, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seldin M, Price A: Application of ancestry informative markers to association studies in European Americans. PLoS Genet 4: e5, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song XW, Di Giovanni V, He N, Wang KR, Ingram A, Rosenblum N, Pei Y: Systems biology of autosomal dominant polycystic kidney disease: Computational identification of gene expression pathways and integrated regulatory networks. Hum Mol Genet 18: 2328–2343, 2009 [DOI] [PubMed] [Google Scholar]
- 19. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: A tool set for whole genome association and population-based linkage analysis. Am J Hum Genet 81: 559–575, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lal M, Song XW, Pluznick J, Di Giovanni V, Rosenblum N, Chauvet V, Gottardi C, Pei Y, Caplan M: Polycystin-1 C-terminal tail associates with β-catenin and inhibits canonical Wnt signaling. Hum Mol Genet 17: 3105–3117, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benzing T, Simons M, Walz G: Wnt signaling in polycystic kidney disease. J Am Soc Nephrol 18: 1389–1398, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Saadi-Kheddouci S, Berrebi D, Romagnolo B, Cluzeaud F, Peuchmaur M, Kahn A, Vandewalle A, Perret C: Early development of polycystic kidney disease in transgenic mice expressing an activated mutant of the beta-catenin gene. Oncogene 20: 5972–5981, 2001 [DOI] [PubMed] [Google Scholar]
- 23. Qian CN, Knol J, Igarashi P, Lin F, Zylstra U, The BT, Williams BO: Cystic renal neoplasia following conditional inactivation of apc in mouse renal tubular epithelium. J Biol Chem 280: 3938–3945, 2005 [DOI] [PubMed] [Google Scholar]
- 24. Niehrs C: Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 25: 7469–7481, 2006 [DOI] [PubMed] [Google Scholar]
- 25. McCarthy M, Abecasis G, Cardon L, Goldstein D, Little J, Ioannidis J, Hirschhorn J: Genome-wide association studies of complex traits: Consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369, 2008 [DOI] [PubMed] [Google Scholar]
- 26. Hirschhorn J, Lohmueller K, Byrne E, Hirschhorn K: A comprehensive review of genetic association studies. Genet Med 4: 45–61, 2002 [DOI] [PubMed] [Google Scholar]
- 27. The Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arking D, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marbán E, O'Donnell CJ, Hirschhorn JN, Kääb S, Spooner PM, Meitinger T, Chakravarti A: A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac re-polarization. Nat Genet 38: 644–651, 2006 [DOI] [PubMed] [Google Scholar]
- 29. Samani N, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, König IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, WTCCC and the Cardiogenics Consortium : Genome-wide association analysis of coronary artery disease. N Engl J Med 357: 443–453, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885, 2007 [DOI] [PubMed] [Google Scholar]
- 31. Plenge R, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, Pe'er I, Burtt NP, Blumenstiel B, DeFelice M, Parkin M, Barry R, Winslow W, Healy C, Graham RR, Neale BM, Izmailova E, Roubenoff R, Parker AN, Glass R, Karlson EW, Maher N, Hafler DA, Lee DM, Seldin MF, Remmers EF, Lee AT, Padyukov L, Alfredsson L, Coblyn J, Weinblatt ME, Gabriel SB, Purcell S, Klareskog L, Gregersen PK, Shadick NA, Daly MJ, Altshuler D: Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet 39: 1477–1482, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Easton D, Pooley K, Dunning A, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, SEARCH collaborators. Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schürmann P, Dörk T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, kConFab AOCS Management Group. Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA: Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hung R, McKay J, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcátová I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P: A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452: 633–637, 2008 [DOI] [PubMed] [Google Scholar]
- 34. Tenesa A, Farrington S, Prendergast J, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, Semple C, Clark AJ, Reid FJ, Smith LA, Kavoussanakis K, Koessler T, Pharoah PD, Buch S, Schafmayer C, Tepel J, Schreiber S, Völzke H, Schmidt CO, Hampe J, Chang-Claude J, Hoffmeister M, Brenner H, Wilkening S, Canzian F, Capella G, Moreno V, Deary IJ, Starr JM, Tomlinson IP, Kemp Z, Howarth K, Carvajal-Carmona L, Webb E, Broderick P, Vijayakrishnan J, Houlston RS, Rennert G, Ballinger D, Rozek L, Gruber SB, Matsuda K, Kidokoro T, Nakamura Y, Zanke BW, Greenwood CM, Rangrej J, Kustra R, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG: Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 40: 631–637, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Manolio T, Brooks L, Collins F: A HapMap harvest of insights into the genetics of common disease. J Clin Invest 118: 1590–1605, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ravine D, Gibson RN, Walker RG, Sheffield LJ, Kincaid-Smith P, Danks DM: Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet 343: 824–827, 1994 [DOI] [PubMed] [Google Scholar]
- 37. Stevens L, Coresh J, Greene T, Levey A: Assessing kidney function: Measured and estimated glomerular filtration rate. N Engl J Med 354: 2473–2483, 2006 [DOI] [PubMed] [Google Scholar]
- 38. Tobit J: Estimation for relationships with limited dependent variables. Econometrica 26: 24–36, 1958 [Google Scholar]
- 39. Liang KY, Zeger SL: Longitudinal data analysis using generalized linear models. Biometrika 73: 13–22, 1986 [Google Scholar]
- 40. Cox DR: Regression models and life-tables. J R Stat Soc 34: 187–220, 1972 [Google Scholar]
- 41. Abecasis GR, Wigginton JE: Handling marker-marker linkage disequilibrium: Pedigree analysis with clustered markers. Am J Hum Genet 77: 754–767, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen WM, Abecasis GR: Family-based association tests for genome-wide association scans. Am J Hum Genet 81: 913–926, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nyholt DR: A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74: 765–769, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J, Ji L: Adjusting multiple testing in multi-locus analyses using the eigenvalues of a correlation matrix. Heredity 95: 221–227, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.