Abstract
RNA helicases are ubiquitous and essential enzymes that function in nearly all aspects of RNA metabolism. The RNA helicase database (www.rnahelicase.org) integrates the wealth of accumulating information on RNA helicases in a readily accessible format. The database is a portal that allows straightforward retrieval of comprehensive information on sequence, structure and on biochemical and cellular functions of all RNA helicases from the most widely used model organisms Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, mouse and human. Also included are RNA helicases from other organisms that are subject to specific investigation. The database is structured according to the most recent helicase classification into helicase superfamilies (SFs) and families, and thus emphasizes phyologenetic relations between RNA helicases as well. Information on individual RNA helicases can be accessed through various browsing routes or through text-based searches of the database.
INTRODUCTION
Nearly all aspects of RNA metabolism involve RNA helicases, enzymes that use ATP to bind or remodel RNA and RNA–protein complexes (1). As one of the largest class of enzymes in RNA metabolism, RNA helicases are encoded by organisms from all kingdoms of life and by many viruses (2). In structure and sequence, RNA helicases are closely related to DNA helicases (3). However, RNA helicases outnumber their DNA bound cousins by a considerable margin and RNA helicases often perform functions substantially different from those attributed to DNA helicases (1–3).
RNA helicases are not only essential for most processes of RNA metabolism including ribosome biogenesis, pre-mRNA splicing and translation initiation (1–3), but also for sensing viral RNAs in the context of the innate immune system and for the biogenesis and function of miRNAs (4,5). Defects or misregulation of certain RNA helicases have been linked to numerous health issues including cancer, neuro-degenerative disorders and infectious diseases (6–9). Given their central biological roles, RNA helicases are subject to intensive ongoing research in diverse fields. To date, more than 7000 articles and roughly 500 reviews have been published on subjects related to RNA helicases. Currently, about 500 publications appear per year, with an upward trajectory.
AIMS AND SCOPE OF THE RNA HELICASE DATABASE
With increasing volume and diversity of the accumulating data on RNA helicases the need arises to integrate the wealth of information in a timely and readily accessible format. To address this issue, we have compiled the RNA helicase database (www.rnahelicase.org). This database is a completely restructured version of the DExH/D protein database from 1999, which covered only a subset of RNA helicases (10). We have now expanded the scope to RNA helicases in general. We approached the design of the RNA helicase database from the view of user who is interested in the fast retrieval of comprehensive information on one or more specific RNA helicases, and in examining phylogenetic relations of RNA helicases to each other. Therefore, the RNA helicase database aims foremost to enable researchers to locate and retrieve comprehensive information about sequence, structure, biochemical and cellular function of RNA helicases. The database provides ready access to sequence, structure, biochemical and cellular function of all RNA helicases from the most widely used model organisms Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, mouse and human. Also included are RNA helicases from other organisms that are subject to specific investigation, e.g. NS3 from hepatitis C virus (11), the P4 RNA packaging motor from Φ12 (12) or RNA helicase A from bovine (13). Focus on these RNA helicases covers the vast majority of published data on RNA helicases (estimated to >95%), while providing an enduring structure for the database. Notwithstanding, the database can be readily expanded to other organisms, if this need arises in the future.
In addition to enabling access to information on individual RNA helicases, the database emphasizes phylogenetic relations between related RNA helicases. As briefly outlined below and in more detail in several recent reviews, RNA helicases are categorized in superfamilies and families, and many features are shared between RNA helicases of a given family (3,14,15). Functions and features of one RNA helicase often provide clues for other, less well-characterized enzymes from the same family (3). However, family relations are often not apparent to investigators outside the RNA helicase field. We therefore believe that emphasizing family relations is particularly useful for database users new to research on RNA helicases.
STRUCTURE OF THE DATABASE
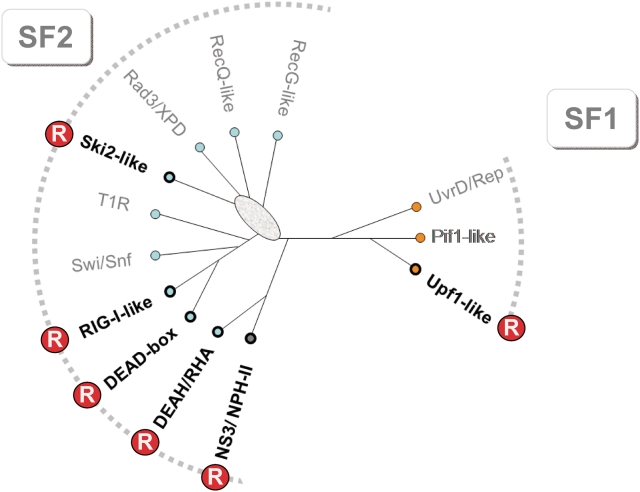
The structure of the RNA helicase database is based on the most recent helicase classification into helicase superfamilies (SFs) and families (3,14,15). According to sequence and protein structures, all RNA and DNA helicases are classified into six SFs (15). SFs 1 and 2 contain non-ring-forming helicases, SFs 3–6 contain ring-forming enzymes. RNA helicases are found in SFs 1–5. All eukaryotic RNA helicases identified to date belong to the non-ring-forming SFs 1 and 2 (3). Ring-forming RNA helicases are found in bacteria and viruses (12,16). SFs 1 and 2 are further divided into families (Figure 1). Although all SF1 and SF2 proteins share extensive structural similarities in the overall fold of their helicase core, the respective families have distinct structure and sequence characteristics that are reflected in specific functional features (3).
Figure 1.
The families of the SF1 and SF2. The cladogram was generated from an structure-aided sequence alignment of all SF1 and 2 proteins in yeast and human, described in (3). The alignment can also be found on the RNA helicase primer page of the database. Branch lengths of the cladogram are not to scale. The oval marks uncertainty in the topology. Families containing RNA helicases are marked with a circled R, the other families contain DNA helicases. (T1R, Type 1 restriction enzymes; RHA, RNA helicase A).
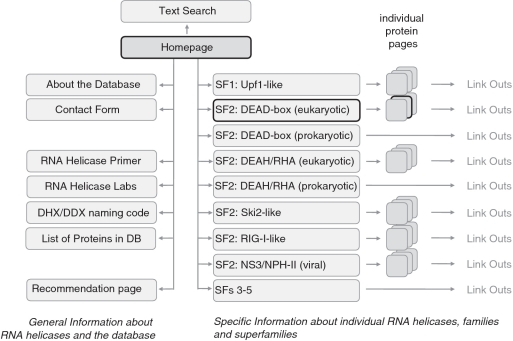
The user enters the database through the homepage (Figure 2). From this page, via the main navigation (Figure 3A), pages for the helicase SFs 3–5, as well as pages for the SF1 and 2 families can be accessed (Figures 2 and 3B). From these pages, one can directly reach ‘individual protein pages’ (Figure 2). These pages contain information on orthologs of a given RNA helicase from S. cerevisiae, C. elegans, D. melanogaster, mouse and human (Figure 3C). DEAD-box and DEAH/RHA helicases from E. coli are listed on separate pages (Figure 2), because there are considerably fewer enzymes than in eukaryotes and because the structure of the individual protein pages, which emphasizes orthologs of multiple organisms, appears not suitable for representing only one protein from E. coli.
Figure 2.
Structure of the RNA helicase database. Gray fields represent database pages, lines show the main connections. Link out indicates links to external sequence databases (see text for details). The bold lines around the Homepage, the SF2: DEAD-box page and one individual DEAD-box protein page mark pages shown as screenshots in Figure 3.
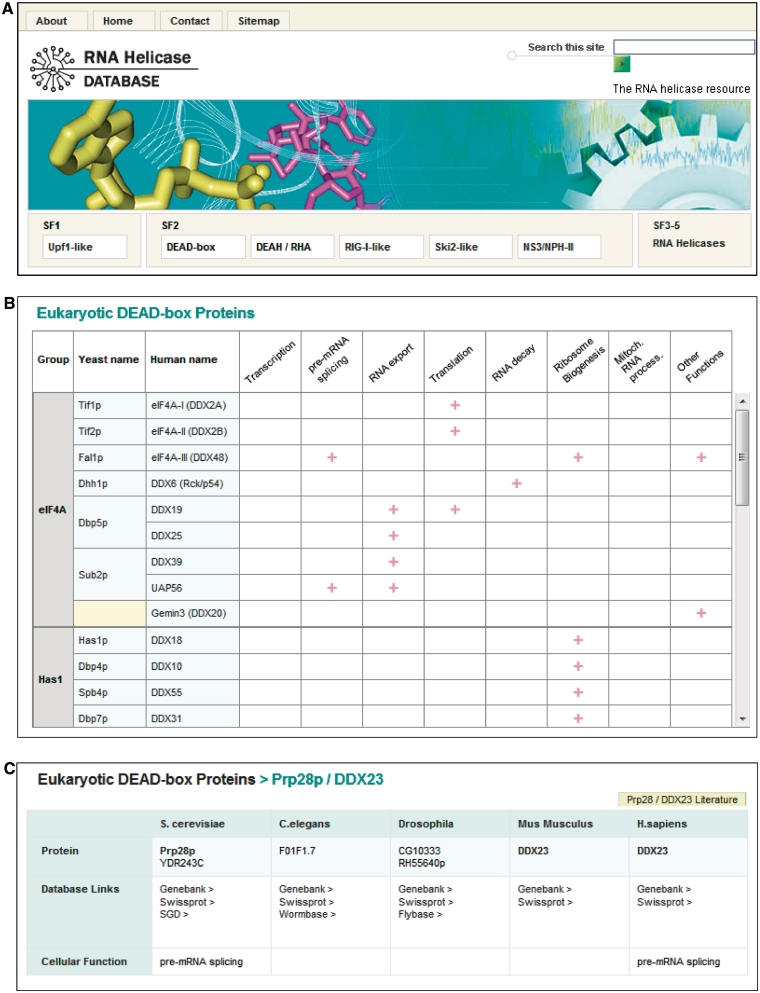
Figure 3.
Screenshots of representative pages. (A) The main navigation is identical on all pages and provides immediate access to the SF3–5 and SF1 and 2 family pages, as well as to the search function. (B) Screenshot of the page for eukaryotic DEAD-box proteins. The pages for individual proteins are accessed through both yeast and human names. Cellular function marks the demonstrated involvement of one ortholog from either organism in the indicated process. (C) Page for an individual protein (Prp28p). Database links connect directly to the specific protein in the sequence databases indicated. Cellular function is only marked if involvement in a specific process has been directly demonstrated for a given protein. The (Prp28/DDX23) literature button provides a direct link to a customized PubMed search for all orthologs listed on the page.
Individual protein pages provide links to various sequence databases with original information about a particular RNA helicase. Links for each protein exist to Genbank and Uniprot, as well as to organism-specific databases SGD (S. cerevisiae), Wormbase (C. elegans) and Flybase (D. melanogaster) (Figure 3C). If structural information is available, respective links to the PDB database are given. The user also has the option to perform a targeted PubMed literature search for the helicase orthologs (Figure 3C). The cellular function of each protein is given only where explicitly tested, although it is generally believed that the cellular functions are largely conserved for orthologs. Finally, a curated alignment of the listed orthologs is provided. This alignment enables the user to identify regions of conservation among orthologs that extend beyond the characteristic helicase motifs.
Individual protein pages were designed based on our roughly decade-long experience with the DExH/D database. Guided by this experience, we chose to directly refer to the large sequence databases for detailed protein information, and not to copy this information into our database. Directly linking to comprehensive, constantly updated sequence databases ensures that information is current without direct intervention from our side.
Further features of the RNA helicase database include full-search ability by text-based queries that are commonplace from the ubiquitous Google search engine (Figure 3A). The search function can be accessed from any page and provides an alternative, yet important route to individual protein pages. A third route of access to individual protein pages is established by a list of proteins in the database (Figure 2). This list shows all RNA helicases in our database, ordered by SF and family and by the organism of origin. Our rationale for incorporating the various routes of access to the individual protein pages is the heterogeneity in the database users, which ranges from researchers looking for updates on information on a helicase that they have long been working on, to those who retrieve the first comprehensive information on a protein they just came across in their research. Finally, the database also contains other information of interest for research on RNA helicases, such as an RNA helicase primer, the naming code for DDX/DHX RNA helicases and a list of laboratories with specific interest in RNA helicases.
FUNDING
Funding for open access charge: National Institutes of Health (GM067700 to E.J.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the many users of previous versions of our database for constructive feedback and valuable suggestions.
REFERENCES
- 1.Jankowsky E. RNA Helicases at work: binding and rearranging. Trends Biochem. Sci. 2010 doi: 10.1016/j.tibs.2010.07.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jankowsky E, Fairman-Williams ME. An introduction to RNA helicases: superfamilies, families, and major themes. In: Jankowsky E, editor. RNA helicases. Vol. 19. London: RSC Biomolecular Sciences; 2010. pp. 1–31. [Google Scholar]
- 3.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- 5.Linder P, Owttrim GW. Plant RNA helicases: linking aberrant and silencing RNA. Trends Plant Sci. 2009;14:344–352. doi: 10.1016/j.tplants.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim. Biophys. Acta. 2004;1704:37–46. doi: 10.1016/j.bbcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Fogel BL, Perlman S. Novel mutations in the senataxin DNA/RNA helicase domain in ataxia with oculomotor apraxia 2. Neurology. 2006;67:2083–2084. doi: 10.1212/01.wnl.0000247661.19601.28. [DOI] [PubMed] [Google Scholar]
- 8.Nelson PT, Keller JN. RNA in brain disease: no longer just “the messenger in the middle”. J. Neuropathol. Exp. Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- 9.Kolb SJ, Sutton S, Schoenberg DR. RNA processing defects associated with diseases of the motor neuron. Muscle Nerve. 2010;41:5–17. doi: 10.1002/mus.21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jankowsky E, Jankowsky A. The DExH/D protein family database. Nucleic Acids Res. 2000;28:333–334. doi: 10.1093/nar/28.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frick DN, Lam AM. Understanding helicases as a means of virus control. Curr. Pharm. Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuma R. Hexameric Viral RNA Helicases. In: Jankowsky E, editor. RNA Helicases. Vol. 19. London: RSC Biomolecular Sciences; 2010. pp. 213–242. [Google Scholar]
- 13.Zhang S, Grosse F. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim. Biophys. Sin. 2004;36:177–183. doi: 10.1093/abbs/36.3.177. [DOI] [PubMed] [Google Scholar]
- 14.Gorbalenya AE, Koonin EV. Helicases: amino acid comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 15.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 16.Rabhi M, Rahmouni R, Boudvillain M. Transcription termination factor rho: a ring-shaped RNA helicase from bacteria. In: Jankowsky E, editor. RNA helicases. Vol. 19. London: RSC Biomolecular Sciences; 2010. pp. 243–271. [Google Scholar]