Abstract
AmoebaDB (http://AmoebaDB.org) and MicrosporidiaDB (http://MicrosporidiaDB.org) are new functional genomic databases serving the amoebozoa and microsporidia research communities, respectively. AmoebaDB contains the genomes of three Entamoeba species (E. dispar, E. invadens and E. histolityca) and microarray expression data for E. histolytica. MicrosporidiaDB contains the genomes of Encephalitozoon cuniculi, E. intestinalis and E. bieneusi. The databases belong to the National Institute of Allergy and Infectious Diseases (NIAID) funded EuPathDB (http://EuPathDB.org) Bioinformatics Resource Center family of integrated databases and assume the same architectural and graphical design as other EuPathDB resources such as PlasmoDB and TriTrypDB. Importantly they utilize the graphical strategy builder that affords a database user the ability to ask complex multi-data-type questions with relative ease and versatility. Genomic scale data can be queried based on BLAST searches, annotation keywords and gene ID searches, GO terms, sequence motifs, protein characteristics, phylogenetic relationships and functional data such as transcript (microarray and EST evidence) and protein expression data. Search strategies can be saved within a user’s profile for future retrieval and may also be shared with other researchers using a unique strategy web address.
INTRODUCTION
Access and effective interrogation of large genomic and functional genomic data sets is challenging but necessary for researchers to mine data generated by others to facilitate their own research programs. The EuPathDB Bioinformatic Resource center (BRC) provides functional genomic databases for the scientific community studying eukaryotic pathogens (1–6). Information regarding all BRCs may be accessed through the BRC portal site (http://www.pathogenportal.net/). Strong emphasis is placed on NIAID’s list of category A–C and (re)emerging pathogens (http://www.niaid.nih.gov/topics/emerging/Pages/list.aspx). Hence, AmoebaDB was established to minimally incorporate genomic data for Entamoeba and Acanthamoeba species, while MicrosporidiaDB is designed to include all genomic data for microsporidial species.
Initial genomic data for AmoebaDB and MicrosporidiaDB were transferred from the previous BRCs that supported these genomes (Pathema for Entamoeba spp. and BioHealthBase for microsporidians) (7,8). In the EuPathDB framework, we have added the ability to integrate functional genomic data with the genome data. To facilitate a smooth launch of these databases and continued successful development, focus was placed on listening to community needs to ensure that these databases provide the resources needed by the scientific communities they serve. Essential to this process are ongoing monthly conference calls hosted by EuPathDB where members of the Amoeba or Microsporidia research communities are invited to call in to provide their insight and knowledge of data sets that should be integrated. Their suggestions help us prioritize tool development and data integration tasks. Both databases were launched in February of 2010 and have since been enhanced with new features and additional data.
EuPathDB database releases occur on a bi-monthly schedule, allowing effective scheduling of development and data integration tasks. This model enables the assignment of specific target release dates for new features and data sets. Data providers are given access to a password-protected development site in advance of a scheduled release so they can approve and suggest changes regarding the representation of their data. Scientists are encouraged to contact EuPathDB (help@eupathdb.org) to discuss their data/experiments in advance of publication in order to ensure release of their data close to publication date. Indeed, in many cases data is approved for release on EuPathDB sites prior to publication and does not jeopardize publication.
This article describes AmoebaDB (http://AmoebaDB.org) and MicrosporidiaDB (http://MicrosporidiaDB.org) with emphasis on how to effectively utilize these resources.
DATA CONTENT IN CURRENT RELEASE OF AmoebaDB
Pathogenic amoebae include both obligate parasites and primarily free-living species, and constitute a leading cause of parasite-induced morbidity and mortality worldwide. Free-living amoebae that cause disease in humans include Acanthamoeba spp., Balamuthia mandrillaris and Naegleria fowleri all of which can lead to various clinical manifestations including central nervous system infections (9). Acanthamoeba spp. are also notorious for causing a debilitating ocular infection and as potential environmental reservoirs for pathogenic bacteria such as Legionella pneumophila (9,10).
Obligate pathogenic amoebae include Entamoeba spp., which constitute a major cause of parasitic illness and death in humans with estimates of close to 100 000 annual deaths (11). Species of Entamoeba that infect humans include the pathogenic E. histolytica, the avirulent E. dispar, E. moshkovskii, E. polecki, Escherichia coli, E. hartmanni and E. gingivalis (12–15). A detailed review of clinical manifestations of amebiasis is provided elsewhere (16). Another species of Entamoeba, E. invadens (parasite of reptiles) is not clinically relevant to humans but is a useful laboratory tool for studying encystation (17).
The current version of AmoebaDB (AmoebaDB 1.1) contains the sequence and annotation of three Entamoeba spp.—E. dispar strain SAW760, E. invadens strain IP1 and E. histolytica strain HM-1:IMSS. The ∼20-Mb genome of E. histolytica is assembled into 1496 scaffolds and contains approximately 8300 gene annotations (18,19). Entamoeba dispar genomic sequence (∼30 Mb) has been assembled into 3312 scaffolds (approximately 8700 annotated genes) (http://www.ncbi.nlm.nih.gov/nuccore/145680688) and E. invadens sequences has been assembled into 1149 scaffolds (approximately 11 500 annotated genes), (http://www.ncbi.nlm.nih.gov/nuccore/AANW00000000).
AmoebaDB 1.1 also contains transcript expression evidence based on expressed sequence tag (EST) data (E. histolytica and E. dispar) obtained from dbEST (http://www.ncbi.nlm.nih.gov/projects/dbEST/) and microarray expression data (E. histolytica) from two studies investigating changes in gene expression during encystation and mouse adaptation (20,21).
DATA CONTENT IN CURRENT RELEASE OF MicrosporidiaDB
Microsporidia are obligate intracellular amitochondriate fungal parasites that infect both vertebrates and invertebrates and are especially common as an insect and fish infection (22). Fourteen microsporidian species infect humans and can lead to severe manifestations in immunocompromised individuals (23). Enterocytozoon bieneusi is by the far the most clinically relevant human microsporidian pathogen, causing severe clinical manifestations in immunocompromised individuals (23).
The current version of MicrosporidiaDB (MicrosporidiaDB 1.1) contains sequence and annotation data for three microsporidian species. Encephalitozoon cuniculi strain GB-M1 has a small 2.9-Mb genome containing 2053 annotated genes, distributed among 11 chromosomes (24). Encephalitozoon intestinalis has 1889 annotated genes, distributed among 11 chromosomes (this sequence was graciously provided by Patrick Keeling and made available prepublication to the community via MicrosporidiaDB). Enterocytozoon bieneusi H348 has an estimated 6-Mb genome with 3806 annotated genes distributed in three chromosomes (25,26). In addition, EST data from dbEST for E. cuniculi are included.
USING AmoebaDB AND MicrosporidiaDB
Genome and gene sequences are subjected to several standard analyses prior to integration in their respective databases. These analyses make several useful searches possible. For example, the sequences can be searched for identification of open reading frames, ESTs that overlap with annotated genes, orthology and synteny between related species and several other searches described in Table 1.
Table 1.
Searches available in AmoebaDB and MicrosporidiaDB
| Search type | Description |
|---|---|
| Text | Genes may be identified based on the presence of a particular word anywhere in the gene record. A user may search all gene record fields or select specific categories such as product name or user comments. |
| Gene IDs | Genes can be identified based on their IDs. This includes current and old IDs. |
| Species | Genes may be identified based on the species they are present in. |
| Genomic location | Genes may be identified based on their exact scaffold, contig or chromosome coordinates. |
| Gene type | Genes may be identified based on whether they are coding or non-coding. |
| Exon count | Genes may be identified based on number of exons. |
| Molecular weight | Genes may be identified based on their calculated molecular weight from the predicted proteins. |
| Isoelectric point | Genes may be identified based on their calculated isoelectric point from the predicted proteins. |
| Predicted signal peptide | Genes may be identified based on whether their predicted protein contains a SignalP predicted signal zpeptide. |
| Transmembrane domain count | Genes may be identified based on the number of predicted transmembrane domains present in their predicted proteins. |
| Protein motif pattern | Genes may be identified based on the presence of a specific amino acid motif. |
| InterPro domain | Genes may be identified based on the presence of InterPro domains in their predicted proteins. |
| BLAST | Genes may be identified using BLAST. |
| EST evidence | Genes may be identified based on whether they also have EST evidence of expression. |
| Microarray evidencea | Genes may be identified based on their expression profile determined from available microarray experiments. |
| GO term | Genes may be identified based on genome ontology terms. |
| EC number | Genes may be identified based on enzyme commission numbers. |
| Orthologs and paralogs | For the identification of orthologs and paralogs of a genes of interest in available species. |
| Orthology phylogenetic profile | Genes may be identified based on whether they have orthologs or paralogs in other organisms. |
aCurrently only available for AmoebaDB.
Starting at the home page
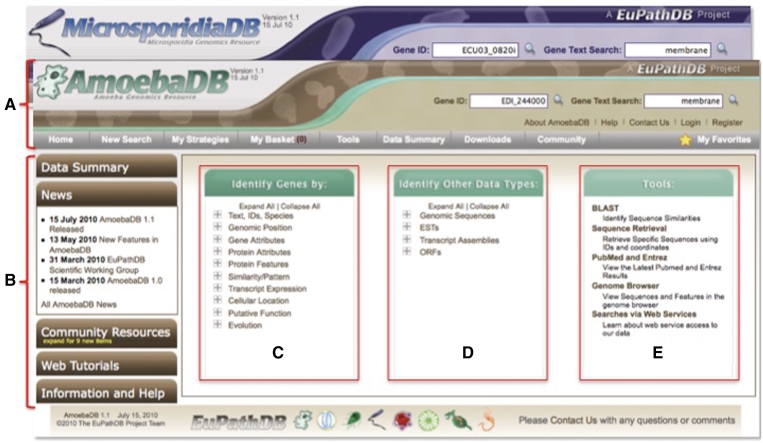
The AmoebaDB and MicrosporidiaDB home pages are virtually identical to all EuPathDB pages, with differences in color, logo and data content only. A visitor to these sites will first notice the home page layout, which has been designed to provide an intuitive and organized venue to access all data and tools. The home page is divided into three main sections (Figure 1); a top banner section, a left-hand information and help section and a central section linking to all searches and tools. The banner section follows a website user on all internal web pages providing quick access back to the home page, gene ID and text searches, ‘contact us’ and login/registration links, and mouse over menus (gray tool bar) providing access to all links in these resources (Figure 1A). The information and help menus on the left (Figure 1B) provide access to a data summary table, database specific news, useful community links and web tutorials. The central section provides links to gene-based searches (Figure 1C), searches against other data types such as genomic sequences, expressed sequence tag data and open reading frames (Figure 1D), and links to tools such as BLAST, the sequence retrieval tool and the genome browser (Figure 1E).
Figure 1.
Screen shot of the AmoebaDB homepage. The layout of the MicrosporidiaDB homepage is exactly the same. (A) The banner section is a constant fixture on all AmoebaDB and MicrosporidiaDB web pages. It contains links to register and login, all searches, tools and information pages, the basket and favorites and ID and text search windows for rapid access from anywhere on the sites. (B) The side panel on the homepage provides expandable tabs that reveal links to a EuPathDB data summary table, community news items, links to community resources, web tutorials and additional information and help links. The central portion of the home page includes all available searches and tools—gene specific searches in (C), searches against other data types such as genomic sequence, open reading frames and ESTs in (D) and tools, such as BLAST, sequence retrieval and the genome browser in (E).
Performing a search
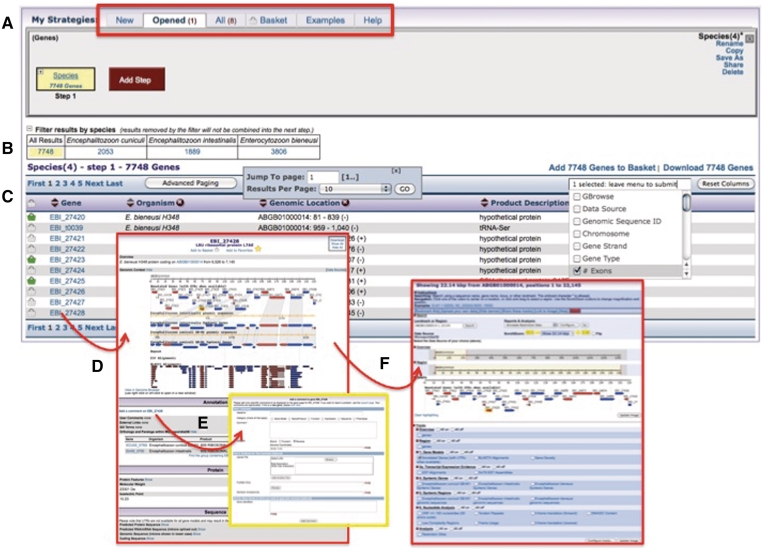
To perform a search in AmoebaDB or MicrosporidiaDB, a user can start with one of the approximately 40 different available searches. These include finding genes based on identifiers, text (including predicted gene product), species, cellular location, BLAST, transcript expression and orthology. Also, genomic sequences, expressed sequence tags (ESTs) and open reading frames can be searched. Table 1 shows a list and description of available gene searches in AmoebaDB and MicrosporidiaDB. After the first search is run, the next page to appear displays a search strategy builder that includes a graphical representation of the search (Figure 2A), a clickable filter table showing the distribution of the results among the species present in the database (Figure 2B) and a summary of the results (Figure 2C). The search strategy section allows a user to access all strategies including ones in their user profile history, items added to their basket (see below), sample strategies and a search strategy-specific help section (tabs highlighted by red rectangle in Figure 2A). In addition, the graphical representation of the search allows a user to quickly see the total number of results or revise the search by clicking on the search name to reveal a popup window with several options.
Figure 2.
AmoebaDB or MicrosporidiaDB results page. Once a user runs a search they are taken to the result summary page that includes a search strategy builder in (A), a filter table showing the distribution of results among related species in (B) and an interactive and configurable table of results in (C). Clicking on the IDs in the results reveals the record page—a gene record page is shown in (D) where a user can view gene-specific information such as synteny and available data, and can add user comments (E) or link to the genome browser to view additional data displayed in the genomic context (F).
The filter table in Figure 2B allows a user to view the distribution of results among the species within the database and specifically view results from each cell in the table by clicking on the numbers in the cells. Importantly, engaging the filter table will also apply the same filter on the search strategy in Figure 2A. Yellow highlighting visually indicates which item in the filter table is currently being viewed (Figure 2B). Results of a search appear in an interactive table (Figure 2C) that allows a user to add and remove columns, sort the results based on items in each column, define advanced paging options, download the results, add results to the user’s basket (see below) and link to gene pages by clicking on gene IDs (Figure 2D). The gene page (Figure 2D) contains gene-centric information, including sections for genomic context (synteny, annotation, neighboring genes, genomic location), annotation, protein features, transcript expression and the gene sequence itself (coding, translated, genomic). The gene page also offers access to the user comment form (described below) (Figure 2E, yellow insert) and to the genome browser (Figure 2F) where additional tracks can be turned on and displayed in the context of a genomic section.
Building a search strategy
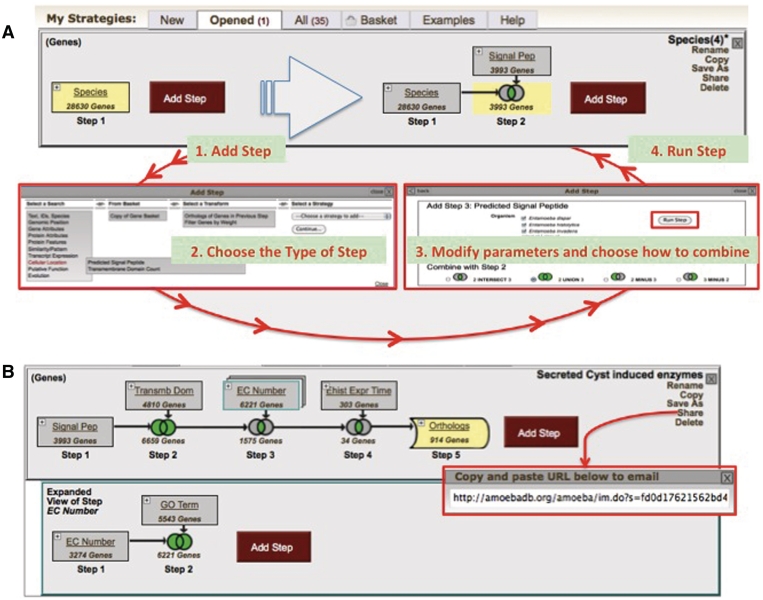
After running a first search (Step 1 in Figure 2A), a user may be interested in refining their results by combining them with other searches. This can be achieved by sequentially adding new searches to grow the strategy horizontally. Figure 3 shows the search strategy cycle: adding steps to a strategy can be done by clicking on the ‘Add Step’ button (1, Figure 3A), a popup window offers the user a set of searches to chose from (2, Figure 3A), once a search is selected a user can modify its parameters and choose how to combine the results with the previous step (3, Figure 3A), to complete the cycle a user runs the step (4, in Figure 3A). The search strategy cycle can be repeated to add additional steps, and each step can be revised, renamed or deleted as needed, steps can be inserted between previously run steps, transformed into orthologs and expanded to generate internal nested strategies.
Figure 3.
A search strategy. (A) The search strategy cycle begins with adding a step to the first search (1), which reveals a popup window with all search options. Once a search type is selected (2), search parameters can be modified and a user chooses how to combine these results with the previous step (3). Clicking on the run step button completes the cycle and expands the strategy horizontally (4). This cycle can be repeated to develop complex search strategies (B). Individual steps in a strategy can be revised, renamed, deleted or transformed into orthologs. Also, the strategy itself can be renamed, copied, saved, deleted and shared with a unique URL (red insert).
An example of a complex multi-step search strategy can be seen in Figure 3B. In this strategy, all Entamoeba cyst-induced secretory enzymes are identified. This is achieved by finding all genes with predicted secretory signal peptides and/or transmembrane domains (Steps 1 and 2, Figure 3B), and an enzyme commission number and/or a metabolic process genome ontology (Step 3 nested, Figure 3B), and are induced in cysts (Step 4, Figure 3B). As a final step, a transformation is applied on the results to identify all Entamoeba orthologs of the results in Step 4 (Step 5, Figure 3B). Several options can be applied to a whole strategy including renaming, copying, saving, deleting and sharing. The latter allows users to email colleagues a unique URL of a strategy of interest, which enables the receiver to open and modify the strategy in their own work space (for example, the strategy in Figure 3B can be accessed here: http://amoebadb.org/amoeba/im.do?s=c63547dbdf9c35bf
ADDITIONAL FEATURES IN AmoebaDB AND MicrosporidiaDB
AmoebaDB and MicrosporidiaDB contain additional useful features.
Favorites
This feature is designed to provide a place where users can bookmark their favorite and frequently accessed genes and to serve as an in silico gene notebook. Adding a gene to favorites can be done by clicking on the star icon underneath the gene product name on gene pages (Figure 2D, yellow star indicates gene has been added to favorites). The favorites page can be quickly accessed from the gray tool bar in the banner (Figure 1A), where genes can be assigned to projects and gene-specific notes added.
Basket
The basket is designed to allow users to cherry pick items including genes, genomic sequences and ESTs for subsequent incorporation into a search strategy. Adding items to the basket can be achieved by clicking on the basket icon found next to gene IDs in the results summary table (Figure 2C, green basket indicates item has been included), clicking on the basket item on any record page, such as the gene record page (Figure 2D, basket icon located under product name), or by clicking on the ‘Add all’ link above the results summary table (Figure 2C). Accessing the basket can be achieved by clicking on the basket link in the gray tool bar in the banner (Figure 1A) or the basket tab in the search strategy builder section (Figures 2A and 3A). Items in the basket can be incorporated (saved) into a search strategy as if they were results of a search (i.e. they can be joined, intersected, etc. with other search results).
Weighted searches
This feature enables a user to assign arbitrary weights to each step in a strategy so that the final list of results are sorted (or ranked) based the sum of all weights in a search strategy. The advantage of using this feature is that items that did not meet all the search criteria are still retrieved and are ranked logically. To explore the outcome of using this feature, compare the search strategy shown in Figure 3B with its weighted counterpart here: http://amoebadb.org/amoeba/im.do?s=fd0d17621562bd43
Running a weighted strategy requires using union set operation to combine results between steps and assigning a weight to each step. As a consequence, the total number of returned results increases dramatically. However, rather than returning a randomly ordered list, the results are sorted based on the sum of their scores (a gene present in each individual step would possess the highest score and would be present in the final list of results if an intersect set operation were used in an unweighted strategy). To further refine this list of results, a user can add a step that filters the results based on a range of weights.
User comments
This feature allows users to provide comments on gene and other record types. The comment form can be accessed from the gene page (Figure 2E, yellow insert). The comment itself may include textual information, references (entering a PubMed ID automatically retrieves the citation), NCBI accession numbers, images and documents and location coordinates. In addition, a comment may be associated with other records in the database using their IDs. Once a comment is submitted it appears instantly on the record page and becomes searchable using the text search tool. This feature enables a user to provide information for the benefit of the community with instantaneous results.
DATA DOWNLOAD AND SEQUENCE RETRIEVAL
All underlying genomic sequence data are available for download from the ‘Data Files’ section accessible from the ‘Download’ section in the gray tool bar (Figure 1A). Data files are organized in folders based on release number and species and include genome annotation in GFF format, various nucleotide and amino acid files in FASTA format and text files including a codon usage table and InterPro features table. In addition to downloading data files in bulk, specific sets of results from searches can be customized and downloaded.
Sequence can be retrieved using the sequence retrieval tool that is accessible from the ‘Tools’ menu on the home page (Figure 1). Specific sequence download coordinates can be defined using this tool and multiple sequences can be downloaded simultaneously. For example, the 500-nt upstream of a set of genes can be downloaded in FASTA format.
FUTURE DIRECTIONS
Both AmoebaDB and MicrosporidiaDB are expected to grow rapidly and dramatically over the next 4 years. This includes genomic sequence data for several additional Entamoeba and Microsporidia species, and genome sequence data for Acanthamoeba spp. Moreover, incorporation of several high-throughput genomic data sets is planned including microarray, proteomics, RNA-Sequence, ChIP-chip and ChIP-sequence and metabolomics. The infrastructure for incorporating many of these data types already exists within EuPathDB databases (i.e. see PlasmoDB, TriTrypDB or ToxoDB). In addition, several genome sequences are planned and already underway (The Centre for Genomic Research at the University of Liverpool and the J. Craig Venter Institute are sequencing several Entamoeba strains and the Broad institute is sequencing microsporidia species).
Community input with annotation is facilitated by user comments, which are forwarded to the sequencing centers. This has already been done in the case of the Toxoplasma sequencing project at JCVI, kinetoplast annotation efforts at GeneDB and will be extended for Entamoeba annotation efforts at JCVI.
FUNDING
National Institute of Allergy and Infectious Diseases (EuPathDB); National Institutes of Health; Department of Health and Human Services; under Contract No. HHSN272200900038C (to D.S.R., C.J.S. and J.C.K.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank members of the Amoeba and Microsporidia research communities for their willingness to share genomic-scale data sets, often prior to publication, and for numerous comments and suggestions that have helped to improve the functionality of AmoebaDB and MicrosporidiaDB. We also thank past and present staff associated with the EuPathDB BRC project, and our research laboratory colleagues whose contributions have facilitated the creation and maintenance of this database resource.
REFERENCES
- 1.Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, et al. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, Rhodes P, Wang S, He CZ, Su Y, et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34:D419–D422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkac LM, Davidsen T, Beck E, Ganapathy A, Caler E, Dodson RJ, Durkin AS, Harkins DM, Lorenzi H, Madupu R, et al. Pathema: a clade-specific bioinformatics resource center for pathogen research. Nucleic Acids Res. 38:D408–D414. doi: 10.1093/nar/gkp850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squires B, Macken C, Garcia-Sastre A, Godbole S, Noronha J, Hunt V, Chang R, Larsen CN, Klem E, Biersack K, et al. BioHealthBase: informatics support in the elucidation of influenza virus host pathogen interactions and virulence. Nucleic Acids Res. 2008;36:D497–D503. doi: 10.1093/nar/gkm905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 10.Harb OS, Gao LY, Abu Kwaik Y. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2000;2:251–265. doi: 10.1046/j.1462-2920.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- 11.Ximenez C, Moran P, Rojas L, Valadez A, Gomez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect. Genet. Evol. 2009;9:1023–1032. doi: 10.1016/j.meegid.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Silberman JD, Clark CG, Diamond LS, Sogin ML. Phylogeny of the genera Entamoeba and Endolimax as deduced from small-subunit ribosomal RNA sequences. Mol. Biol. Evol. 1999;16:1740–1751. doi: 10.1093/oxfordjournals.molbev.a026086. [DOI] [PubMed] [Google Scholar]
- 13.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin. Microbiol. Rev. 2007;20:511–532. doi: 10.1128/CMR.00004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond LS, Clark CG. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J. Eukaryot. Microbiol. 1993;40:340–344. doi: 10.1111/j.1550-7408.1993.tb04926.x. [DOI] [PubMed] [Google Scholar]
- 15.Haque R, Petri WA., Jr Diagnosis of amebiasis in Bangladesh. Arch. Med. Res. 2006;37:273–276. doi: 10.1016/j.arcmed.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N. Engl. J. Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 17.Vazquezdelara-Cisneros LG, Arroyo-Begovich A. Induction of encystation of Entamoeba invadens by removal of glucose from the culture medium. J. Parasitol. 1984;70:629–633. [PubMed] [Google Scholar]
- 18.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 19.Lorenzi HA, Puiu D, Miller JR, Brinkac LM, Amedeo P, Hall N, Caler EV. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl. Trop. Dis. 4:e716. doi: 10.1371/journal.pntd.0000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 21.Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, Evans C, Martino-Catt S, Baba DJ, Stroup S, et al. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Keeling PJ, Fast NM. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 23.Didier ES, Weiss LM. Microsporidiosis: current status. Curr. Opin. Infect. Dis. 2006;19:485–492. doi: 10.1097/01.qco.0000244055.46382.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 25.Akiyoshi DE, Morrison HG, Lei S, Feng X, Zhang Q, Corradi N, Mayanja H, Tumwine JK, Keeling PJ, Weiss LM, et al. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 2009;5:e1000261. doi: 10.1371/journal.ppat.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corradi N, Akiyoshi DE, Morrison HG, Feng X, Weiss LM, Tzipori S, Keeling PJ. Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi. PLoS ONE. 2007;2:e1277. doi: 10.1371/journal.pone.0001277. [DOI] [PMC free article] [PubMed] [Google Scholar]