Abstract
We updated the tRNADB-CE by analyzing 939 complete and 1301 draft genomes of prokaryotes and eukaryotes, 171 complete virus genomes, 121 complete chloroplast genomes and approximately 230 million sequences obtained by metagenome analyses of 210 environmental samples. The 287 102 tRNA genes in total, and thus two times of the tRNA genes compiled previously, are compiled, in which sequence information, clover-leaf structure and results of sequence similarity and oligonucleotide-pattern search can be browsed. In order to pool collective knowledge with help from any experts in the tRNA research field, we included a column to which comments can be added on each tRNA gene. By compiling tRNAs of known prokaryotes with identical sequences, we found high phylogenetic preservation of tRNA sequences, especially at a phylum level. Furthermore, a large number of tRNAs obtained by metagenome analyses of environmental samples had sequences identical to those found in known prokaryotes. The identical sequence group, therefore, can be used as phylogenetic markers to clarify the microbial community structure of an ecosystem. The updated tRNADB-CE provided functions, with which users can obtain the phylotype-specific markers (e.g. genus-specific markers) by themselves and clarify microbial community structures of ecosystems in detail. tRNADB-CE can be accessed freely at http://trna.nagahama-i-bio.ac.jp.
INTRODUCTION
More than 99% of microorganisms that inhabit natural environments are difficult to culture under laboratory conditions. Metagenomic analyses of mixed genome samples have been developed (1–3) to explore the enormous number of novel genome resources. In accord with the remarkable progress of DNA-sequencing technology, a vast quantity of metagenomic sequences obtained from a wide variety of environmental samples have been decoded and released from DDBJ/EMBL/GenBank. Because a significant portion of environmental DNA sequences is derived from unculturable microbes, we can acquire new knowledge of tRNA sequences from novel genomes. The 154 455 tRNA genes found in the metagenomic sequences were included in the tRNADB-CE. When we focused on a group of tRNAs with an identical sequence, we found tRNAs found only in a particular lineage of phylogenetic groups. Notably, such phylotype-specific tRNA sequences were found in many species-unknown genomic fragments obtained by metagenome analyses of environmental samples. This shows that tRNA is a good phylogenetic marker for discovering phylotype composition and microbial community structure in an environmental ecosystem.
MATERIALS AND METHODS
The following sources of DNA sequences were used: the complete genomes of 927 prokaryotes and of 171 viruses released by Genome Information Broker (GIB, http://gib.genes.nig.ac.jp/) and Genome Information Broker for Viruses (GIB-V, http://gib-v.genes.nig.ac.jp/) of DDBJ up to March 2009; the complete genomes of 121 chloroplasts released by Organelle Genome Database (GOBASE, http://gobase.bcm.umontreal.ca/) up to March 2009; the draft genome sequences of 1301 prokaryotes released by WGS division of DDBJ/EMBL/GenBank up to August 2009; the complete genomes of 12 eukaryotes and the 17 million metagenomic sequences released by DDBJ/EMBL/GenBank up to March 2010; and the 217 million metagenomic sequences obtained using a next-generation sequencer and released by Sequence Read Achieve (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra/) in NCBI up to March 2010.
RESULTS AND DISCUSSION
Update of registered tRNA genes and a new function for organizing collected knowledge
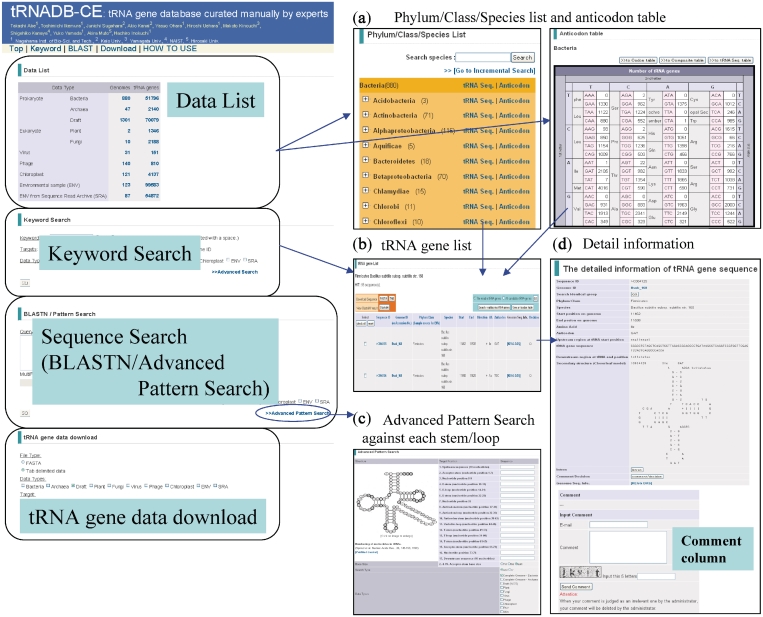
The 287 102 tRNA genes in total (53 936, 70 079, 961, 3534, 4137 and 154 455 genes from 927 complete prokaryote genomes, 1301 draft prokaryote genomes, 171 complete virus, 12 complete eukaryote genomes, 121 complete chloroplast genomes and 210 metagenomic samples, respectively) were registered in the updated tRNADB-CE. This was two times as many tRNA genes as registered in the previous version (4). This exhaustive search for tRNA genes was performed by running three computer programs used for tRNA gene search, tRNAscan-SE (5), ARAGORN (6) and tRNAfinder (7) in combination. This method should enhance the completeness and accuracy of prediction since the programs’ algorithms are partially different and render somewhat different results. The tRNA genes found accordantly by all three programs were stored in tRNADB-CE without further checks. Then, the residual, discordant cases (∼4% of the total of tRNA gene candidates), except for those found in metagenomic sequences, were checked, manually by three experts (Y.Y., A.M. and H.I.) in the tRNA experimental field and were classified into three categories: (i) reliable tRNA genes, (ii) not tRNA genes and (iii) ambiguous cases. Users can browse or download either the reliable tRNA genes or all candidate genes by choosing ‘The reliable tRNA genes’ or ‘All candidate genes’. Criteria used for this manual evaluation were described in detail previously (4). The tRNA genes of Archaea obtained from SPLITSdb (8) were included in this new version. Basic functions of the database were described previously in detail and briefly in Figure 1.
Figure 1.
Basic functions of tRNADB-CE. (a) Phylum/class/species list and anticodon table, (b) tRNA gene list, (c) Advanced pattern search against each stem/loop and (d) Detail information.
To aim at establishing a very reliable database utilizing collective knowledge in the various experimental fields of tRNA research, we developed a new function for including comments on each of the registered tRNA genes on ‘the detailed information of tRNA gene sequence page’ (Figure1d). User can add comments by entering e-mail address and password, while we reserve the right to remove irrelevant comments. We hope that the accumulation of user comments will provide annotation of higher quality and that this database will become an information sharing system in the tRNA gene community.
Identical sequence groups and their use as phylogenetic markers for environmental metagenomic sequences
When we conducted the clustering of 124 015 sequences of tRNA genes, except for the 3′-CCA terminal sequence, from prokaryotic genomes by sequence alignment using the CD-HIT (9), we found high phylogenetic preservation of tRNA genes: i.e. a particular tRNA sequence was found only in a particular lineage of phylogenetic groups. We designated here the tRNA group with an identical sequence as ‘Identical Sequence Group: ISG’ (Figure 2a) and listed the numbers of ISGs for each phylotype (Figure 2b) and for each anticodon (Figure 2c). The tRNAs with one anticodon type were classified and listed according to ISG along with the phylotype information of each tRNA (Figure 2d), and therefore, the range of phylotypes found for each ISG could be examined. If we focused on ISGs composed of more than five sequences, 97.1% of ISGs were conserved at a phylum level, showing most tRNAs to be good phylogenetic markers at least at a phylum level. The ISGs could provide a strategy to select reliable phylogenetic markers. At the genus level, ∼60% of ISGs were conserved, showing there may exist good genus-specific markers. By combining the data provided by this database with other detail knowledge of a particular tRNA obtained by experiments or from literature, users may get useful phylogenetic markers (e.g. genus-specific markers) by themselves.
Figure 2.
List and search for (a) ISG, (b) ISGs for each phylotype, (c) ISGs for each anticodon, (d) ISG tRNAs for one anticodon and (e) ISGs found in an environmental sample.
Interestingly, among 154 455 tRNA genes found in metagenomic sequences derived from environmental samples, 35 739 tRNA genes (23%) were identical in sequence to genes from known prokaryotes. Using tRNAs found in an environment sample that were assigned to ISGs, we could predict microbial community structures in an environmental ecosystem at least at a phylum level (Figure 2e). The database has also a function to search for sequences with 97 or 95% sequence identify (2- or 3-nt difference, respectively) (Figure 2a). By using tools in the database and the specific markers found by users (e.g. genus-specific markers), users can independently clarify microbial populations in an ecosystem. This strategy can be applied even to data of short sequences obtained using next-generation sequencers, such as SRA in NCBI. In metagenomic analysis by a next-generation sequencer where the length of sequences obtained is short, phylogenetic characterization of the short sequences was particularly difficult using existing methods, except for sequences derived from dominant species or sequences unambiguously mapped on a known sequenced genome. Because tRNA genes are searchable even from short genomic fragments of around 100 bases, tRNA genes should become one of the most effective means for identifying the microbial populations in an ecosystem when analyzing a vast number of metagenomic sequences obtained by next-generation sequencers.
ACCESS TO THE DATABASE
tRNADB-CE can be accessed freely from http://trna.nagahama-i-bio.ac.jp.
FUNDING
Integrated Database Project and a Grant-in-Aid for Publication of Scientific Research Results from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Funding for open access charge: Integrated Database Project from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delong EF, Karl DM. Genomic perspectives in microbial oceanography. Nature. 2005;437:336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- 3.Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model commuities to study evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 4.Abe T, Ikemura T, Ohara Y, Uehara H, Kinouchi M, Kanaya S, Yamada Y, Muto A, Inokuchi H. tRNADB-CE: tRNA gene database curated manually by experts. Nucleic Acids Res. 2009;37:D163–D168. doi: 10.1093/nar/gkn692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinouchi M, Kurokawa K. tRNAfinder: A software system to find all tRNA genes in the DNA sequence based on the cloverleaf secondary structure. J. Comput. Aided Chem. 2006;7:116–126. [Google Scholar]
- 8.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]