Abstract
The GPCRDB is a Molecular Class-Specific Information System (MCSIS) that collects, combines, validates and disseminates large amounts of heterogeneous data on G protein-coupled receptors (GPCRs). The GPCRDB contains experimental data on sequences, ligand-binding constants, mutations and oligomers, as well as many different types of computationally derived data such as multiple sequence alignments and homology models. The GPCRDB provides access to the data via a number of different access methods. It offers visualization and analysis tools, and a number of query systems. The data is updated automatically on a monthly basis. The GPCRDB can be found online at http://www.gpcr.org/7tm/.
INTRODUCTION
G protein-coupled receptors (GPCRs) constitute a large family of cell surface receptors. They regulate a wide range of cellular processes, including the senses of taste, smell and vision, and control a myriad of intracellular signaling systems in response to external stimuli. GPCRs are a major target for the pharmaceutical industry as is reflected by the fact that more than a quarter of all FDA approved drugs act on a GPCR (1). GPCRs are arguably one of the most-researched classes of proteins, but despite intensive academic and industrial research efforts over the past three decades, little is known about the structural basis of GPCR function. From about 350 genes that code for the non-olfactorial receptors in the human species (2), only about 30 are truly validated therapeutic targets (3), indicating this family’s immense potential for future drug development. The fact that GPCRs can form homo-oligomeric and hetero-oligomeric complexes (4) has created a lot of new challenges and opportunities in the rational drug design process. In addition, a number of high-resolution crystal structures recently became available, providing new insights in receptor structure and function and giving the GPCR field a big stimulus.
Researchers who focus on one particular protein or a class of proteins are confronted with the fact that both the number and the size of databases are expanding at an ever-increasing pace. Although many databases like PDB (5), UniProtKB (6), KEGG (7), EMBL (8), GenBank (9), etc. are invaluable for their research, for the average wet-lab scientist these databases are less suitable for gathering, integrating, and updating different types of data in an easy and efficient manner. Studies that involve carrying over information from one protein to the other seem simple at a first glance, however, the amount of data that needs to be collected from heterogeneous sources, converted to syntactic and semantic homogeneity, validated, curated, stored and indexed, is enormous.
The GPCRDB is a data source that holds a large amount of heterogeneous data in a well-organized and easily accessible form. This data is validated, internally consistent, and updated regularly. In addition to being a one-stop GPCR resource, the data in the GPCRDB facilitates inferring new information using a wide spectrum of bioinformatics techniques. The GPCRDB is a paradigm for MCSIS technology (10,11).
NEW FEATURES
The previous release of the GPCRDB (12) was almost entirely a static website, neither offering much dynamic content, nor possibilities for complex interactions or the use of computational tools. We addressed this problem by rewriting the entire system. The use of new tools and modern-day e-Science technologies has resulted in improved flexibility and greater user-friendliness. We have updated the methods for harvesting GPCR sequences, expanded the number of data types available, and added new tools and services to the GPCRDB. Nearly all of the functionality that is offered through the web interface is also available in the form of web services. This allows for the easy integration of the GPCRDB in custom built tools and scripts or in workflow management tools such as Taverna (13) and Pipeline Pilot (http://www.accelrys.com/products/pipeline-pilot/). All pages now offer extensive context-sensitive help functionality, explaining what kinds of data are displayed and how to use the available interactive functionalities such as searching and computational tools.
DATA CONTENT
The contents of the GPCRDB can be categorized in three classes: primary, secondary and tertiary data. Sequence data, ligand-binding constants, mutant information, structural data and oligomer interactions make up the experimentally determined primary data. Data types such as multiple sequence alignments (MSAs), homology models, correlation patterns and entropy-variability data are inferred from these primary data, and fall in the category of secondary, or computationally derived data. Curator provided interpretations and other user help facilities make up the tertiary data category. Table 1 shows a few vital statistics about the volume of the data content of the GPCRDB.
Table 1.
Statistics for the September 2010 release of the GPCRDB
| Sequences | 27 045 |
| Families (and MSA) | 1270 |
| Mutations | 7703 |
| Ligand-binding data | 12 086 |
| Protein structures | 195 |
| Homology models | 22 616 |
| Residues | 11 290 993 |
| Species | 1521 |
| Oligomers | 115 |
PRIMARY DATA
Sequences
GPCR sequences are extracted from NCBI’s NR database, which is a non-redundant protein sequence database with entries from a set of sequence repositories that include GenBank CDS translations, UniProtKB and PDB. GPCR sequences are selected by classifying them against a database of Hidden Markov Models (HMMs). For each of the protein families in the GPCRDB a HMM is available. These HMMs are created from MSA of the previous GPCRDB release. HMM files are created with the HMMER software package (http://hmmer.wustl.edu/).
As a first step in harvesting GPCR sequences we perform a BLAST search with all the HMM consensus sequences against the NR database. By using a very relaxed cut-off value we collect a large number of resulting hits, including many false positives. This step is necessary to limit the amount of sequences that will be used for the actual classification while ensuring a minimal false-negative rate. This step reduced the search space (for the September 2010 GPCRDB release) from about 11 million sequences to about 80 000. These hits are scored against the collection of HMMs to place them in the correct family or discard them as not being a GPCR sequence. Additional filter steps are applied such as filtering out sequence fragments and sequences that contain ambiguous amino acid characters, resulting in a final set of about thirty thousand sequences. The corresponding database entries of the selected sequences are retrieved with MRS (14) and additional data such as gene names and species information is extracted and stored. The GPCRDB holds for each sequence one principal access page. Figure 1 shows an example of such a page.
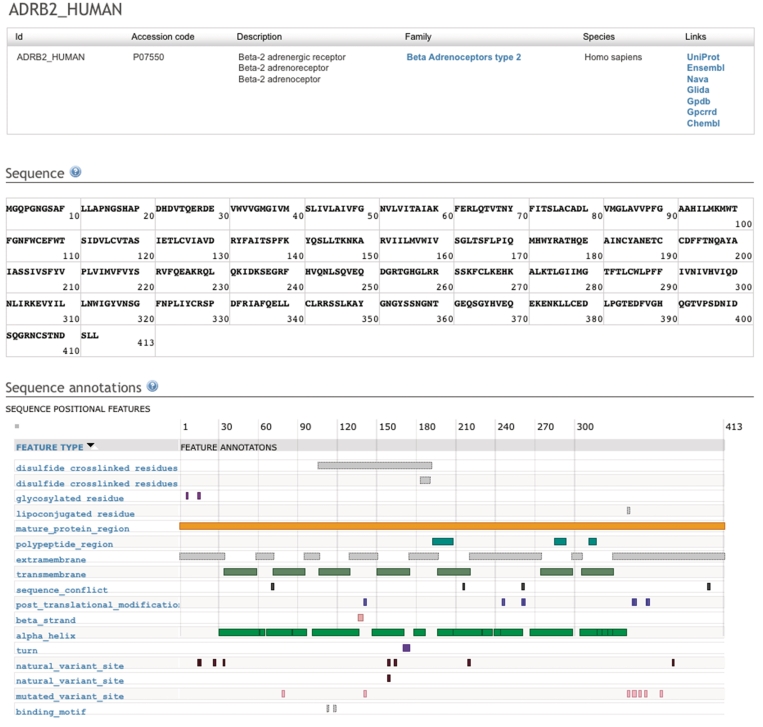
Figure 1.
Screenshot of the principal protein sequence page of the human beta-2 adrenoceptor. The top table contains details about the protein record and hyperlinks to the protein family browsing page and other data sources that contain information about this protein. The middle table holds the sequence in which each amino acid is linked to its own residue page. The bottom table holds annotations that are obtained in real time using the DAS (15) system.
The protein detail page (Figure 1) contains a panel that visualizes sequence annotations such as helix boundaries, cysteine bridges and glycosylation sites. These annotations are loaded in real time using the DAS distributed annotation system (15,16) and are visualized by Dasty2 (17), resulting in always up-to-date annotations. We use the UniProt DAS server to retrieve sequence annotations.
Ligand-binding data
Ligand-binding constants are available for a large number of GPCRs and are obtained from various sources. For each GPCR we provide links, if possible, to the ChEMBL (18) and GLIDA (19) databases. In addition, ligand-binding information that is obtained from collections from Seeman (20) and Organon (21) is available. Since ligand-binding data is very hard to obtain from the literature we encourage academic and industrial researchers to submit their ligand-binding data to the GPCRDB in order to make this data accessible to the scientific community.
Mutations
The GPCRDB contains a large number of well-annotated mutations obtained from different sources. We have two sets of mutations that were manually extracted from literature. Mutant data from the tinyGRAP database (22) contains references to scientific literature describing point mutations as well as insertions, deletions and chimeric receptors. A collection of in-house manually extracted mutant data contains a few thousand point mutations and the effects of those mutations on the function of the receptor. We have extracted sentences from the papers that qualitatively describe the effects of these mutation and we have extracted quantitative data such as effects on ligand binding, expression, activation or constitutive activity.
In addition to the two manually curated data sets we also have a large body of mutations that were extracted from the literature by the software package MuteXt (23). A sentence describing the effects of the mutations is available for all mutations extracted by MuteXt.
Structures
Structures are obtained from the PDB. Links to structures that were re-refined in the PDB_REDO project (24) are included. We provide manually ‘cleaned’ monomers of the major GPCR PDB files that have been prepared for easy casual use by the life sciences community.
Oligomers
GPCR oligomerization has been an area of interest and controversy for many years. Recently there has been increasing evidence that both homo-dimers and hetero-dimers play a crucial role in GPCR signaling (25–27). The GPCR-OKB (28) is a database that stores manually extracted computational and experimental information about GPCR oligomerization. Lists of protomers, experimental details and, where available, inferred oligomer interaction sites are available for all oligomers. This data has been fully integrated in the GPCRDB, making the information about both GPCR protomers and oligomers readily available.
SECONDARY DATA
Multiple sequence alignments
MSAs are available for all families. MSAs are generated with WHAT IF (29) for all GPCR sub-families using hand-optimized sub-family specific profiles. Position-specific annotations such as secondary structure information and generalized residue numbers are stored in the profiles and are incorporated in the alignments. The general residue numbers are relative to the arbitrarily selected numbers for very conserved residues and motifs such as the well-known E/DRY and NPXXY motifs. Using a profile to align a GPCR sub-family allows for the mapping of the general residue numbers on the sequences that are being aligned. The result is that the residues in the TM domains, helix VIII and sections of the loops are labeled with a general residue number.
For creating alignments of parent GPCR families we make use of these general residue numbers. For all the GPCRs that are being aligned we select all the general residue positions that the sub-families have in common and create the alignment by listing, for each sequence, the residues at the selected positions. GPCR parent family alignments are thus not built using standard alignment algorithms but are created by selecting residues that are likely to share the same position in the 3D structure.
Homology models
Despite the recent publication of a number of GPCR structures the amount of structural information on GPCRs is still very limited. We have built structure models of all Class A receptors to at least partially fill this gap. Information extracted from the profile-based MSA is used to generate high-quality sequence–structure alignments between GPCR sequences and a number of experimentally determined structures. Based on these alignments homology models of all GPCRs have been automatically created and will be automatically updated as more sequences become available. Template structures are selected based on sequence identity and a number of structure quality criteria. The homology models are created with WHAT IF and YASARA (30). Models will be automatically replaced when new structures become available that are better templates, with better being defined as either being solved with higher resolution data, or as having a higher percentage sequence identity.
Correlated mutation analyses
Correlated mutation analysis (CMA) is a technique that can find pairs of residues that remain conserved or mutate in tandem during evolution. Residues that show correlated behaviour in MSA are likely to be functionally related, and networks of those correlating residues indicate functional groups (31). The rationale behind this analysis is that when a mutation occurs at a functionally important site, the protein either becomes functionally impaired or may acquire its original or a different function due to compensatory mutations at one or more other positions. Correlation scores are available in a number of different formats for all GPCR (sub)families.
Entropy and variability data
The amount of entropy and variability that is observed for a certain position in multiple sequence alignment tells something about the types of pressures exerted on that position during evolution (32–34). Entropy-variability data is available for each multiple sequence alignment in the GPCRDB. We offer this data in tabular from, entropy-variability plots (35), and more advanced subfamily specific two-entropy plots (manuscript in preparation) based on the original method described by Ye et al. (36) (Figure 2).
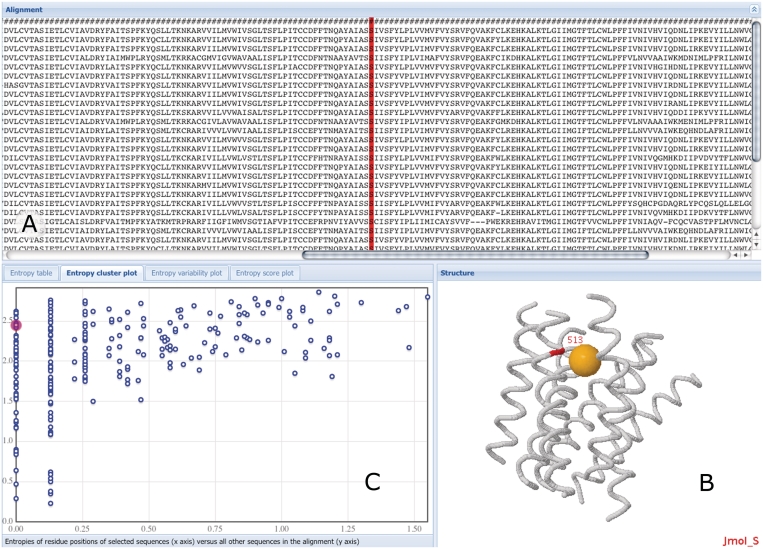
Figure 2.
Screenshot of the interactive entropy and variability page. The multiple sequence alignment is shown in (A). Residues are interlinked in all page elements, clicking results in highlighted selections. In (B), the approximate location of this position in the 3D model of the transmembrane domain of class A GPCRs is shown in red and is annotated with general residue number information. The orange ball in the structure model indicates the approximate location of the assumed binding site for low molecular weight compounds of class A GPCRs. In (C) the user can choose among four display modes that describe the entropy and variability of all positions in the alignment; shown is the entropy cluster variant.
TERTIARY DATA
Residue annotations
Residues in the GPCRDB are labelled with the original Oliveira et al. (37) numbering scheme as well as with residue numbers from the more recent Ballesteros–Weinstein scheme. Use of these general residue numbers allows for easy transfer of information between proteins. General residue numbers are available for all residues within conserved structure elements. These include all transmembrane helices, helix VIII and a few sections in the loops. For each residue with a general residue number a short description of its properties and interactions is available. These descriptions are based on a manual analysis of the currently available crystal structures.
Cytoscape networks
The GPCRDB provides cytoscape (38) network files for all GPCR families. These network files contain the proteins of a family with distances calculated from the family alignments. For all proteins the protein family information, species names and the amino acid types for all the residues annotated with a general residue number are available as attributes. This allows for complex analyses, such as coloring proteins by amino acids at a certain residue position to compare i.e. species or sub-type specific differences.
Mutation predictions
For all positions for which a general residue number is available we have investigated the most likely effects of mutations at these positions. Short fragments of text have been created that explain for each of these positions the likely effects of the mutation on structural and functional levels. References to key papers in which experimental evidence for these effects is available are included in the fragments. Examples of such effects are the loss of ligand-binding affinity when mutating a residue in the ligand-binding pocket, the loss of G protein binding when mutating residues at the G protein-binding interface, and increased constitutive activity when residues are mutated a the interface between helix III and VI.
Workflows
We have created a number of Taverna workflows that use the web services of the GPCRDB as a starting point for users who want to programmatically access the GPCRDB. Workflows are available that use the GPCRDB BLAST service, create custom-built alignments and retrieve several different data types. The workflows and documentation are available via the myExperiment web portal (39) and are tagged with ‘GPCRDB’. We encourage researchers to share their own workflows via the myExperiment portal.
DATA ACCESS
The GPCRDB provides fast and easy access to all its data and information. The GPCRDB does not merely lists available data, but all data types are fully integrated. For example, mutations are accessible via the protein detail pages but can also be found at the residue level. The same holds true for oligomer interaction interfaces, where details about these interactions are available via the oligomer pages, but also via the protein detail pages of interacting members, as well as via the pages of residues that are reported in the interaction. This tight data integration makes it a very efficient resource to use. The GPCRDB’s user interface allows the user to easily navigate from one data type to another and often suggests multiple routes to explore the data, thereby hopefully generating ideas and questions while the user navigates the system.
The four fundamental facilities to be provided by information systems are browsing, retrieval, query and inferencing. These four types of access have been an integral part of the GPCRDB set-up from the beginning. The total redesign of the GPCRDB that has taken place the past few years has allowed us to add novel access facilities in all these four categories.
Browsing
The main way to access the data is via a hierarchical list of GPCR families, which is based on the pharmacological classification of GPCRs (40) (Figure 3). Users can traverse the GPCR family tree and view or download the data for a selected family. Available data types include MSAs, entropy-variability analyses and lists of family members. Alignments can be viewed in multiple ways. In addition to the classic HTML view, the GPCRDB offers an interactive multiple sequence alignment viewer [JalView (41)], that can show additional information about the MSAs, supports a number of viewing and sorting options, and that can be used to generate phylogenetic trees. Residues for which mutation data is available are hyperlinked in the alignments to pages that contain more details about those mutations.
The pages that display data on individual proteins (Figure 1) contain a large amount of data and links to other data sources. Table 2 lists the remote databases that have been indexed in the GPCRDB. Some of these remote data are actually most easily queried via the GPCRDB.
Table 2.
Non-GPCRDB data facilities that can be found through the GPCRDB
| Database | Type of data | Address |
|---|---|---|
| GPCR-OKB (GPCR oligomerization knowledge base) | Dimer information | http://data.gpcr-okb.org/gpcr-okb/ |
| GPCRRD (GPCR restraint database) | Modeling restraints | http://zhanglab.ccmb.med.umich.edu/GPCRRD/ |
| GLIDA (GPCR ligand database) | Ligand data | http://pharminfo.pharm.kyoto-u.ac.jp/services/glida/ |
| gpDB (G protein database) | G Protein data | http://biophysics.biol.uoa.gr/gpDB/ |
| UniProt | Protein information | http://www.uniprot.org/ |
| NAVA | Sequence Variants | http://nava.liacs.nl/ |
| ChEMBL | Ligand data | http://www.ebi.ac.uk/chembl/ |
| Ensembl | Genomic information and annotations | http://www.ensembl.org |
On the protein detail page the sequence is displayed and each residue is hyperlinked to its individual page where additional information is listed about that specific residue; residue numbers in multiple numbering schemes, available mutation data for that specific residue and mutations at equivalent positions, and reported oligomeric interactions.
Snake plots are available for all proteins in the GPCRDB (Figure 4). Residues for which mutations are available are hyperlinked from the snake plots to pages that contain details about those mutations.
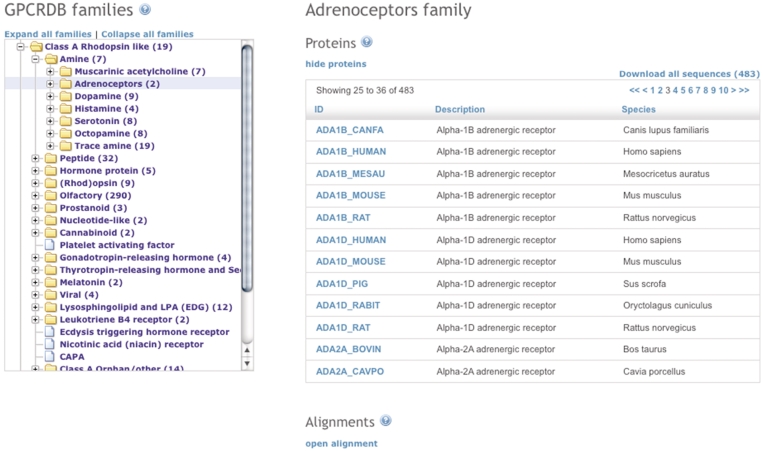
Figure 3.
Screenshot of the GPCR family page. The GPCR family tree is shown on the left with the amine sub-family expanded. On the right-hand side the data for the selected family (adrenoceptors) is shown.
Other data types such as mutations, ligand-binding constants and information about oligomerization states are displayed when available and links are provided to pages that contain more detailed information about those data (Figure 5).
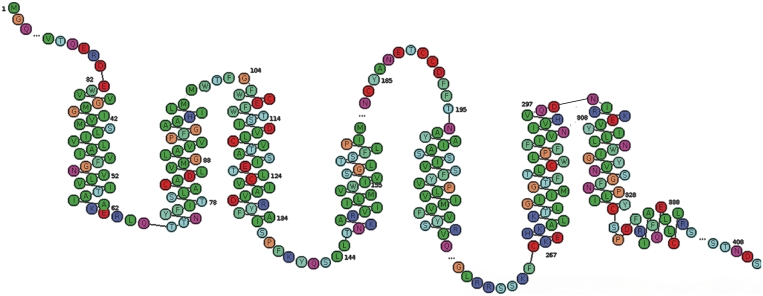
Figure 4.
Snake plot of the human β2 adrenoceptor.
The pages with mutation details contain links to the scientific literature and if available in that literature, the qualitative and quantitative data on the effects of the mutations is displayed. The oligomer detail page contains links to the GPCR-OKB and to the individual protomers in the GPCRDB. If certain residues are involved in the oligomer interaction, hyperlinks to individual residue pages are available.
Retrieval
Data can be retrieved via the web pages and via the web services. The web services offer very extensive retrieval possibilities, together allowing for the retrieval of all data types present in the GPCRDB. Subsets have been created for frequently requested data sets such as all human sequences and the human non-olfactory sequences. Protein family alignments can be downloaded in a number of different formats. Sequences, structures, ligand-binding data and mutations can be downloaded from the protein detail pages. After querying the GPCRDB via the web pages, query result sets can be downloaded in FASTA format. A complete copy of the GPCRDB is freely available for in-house usage by academic and industrial researchers alike.
Query
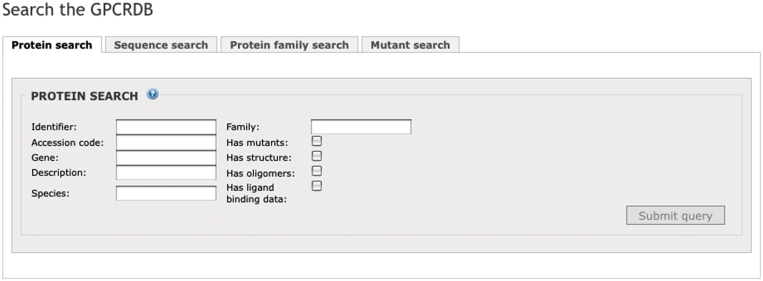
Users can query the GPCRDB via a number of different search pages. Identifiers, genes, species, descriptions and protein family names can be used to search for GPCRs (Figure 6). There are a number of filters available to limit the search results. Users can indicate whether only GPCRs should be shown for which mutations, structures, oligomers, or ligand-binding data are available.
Figure 5.
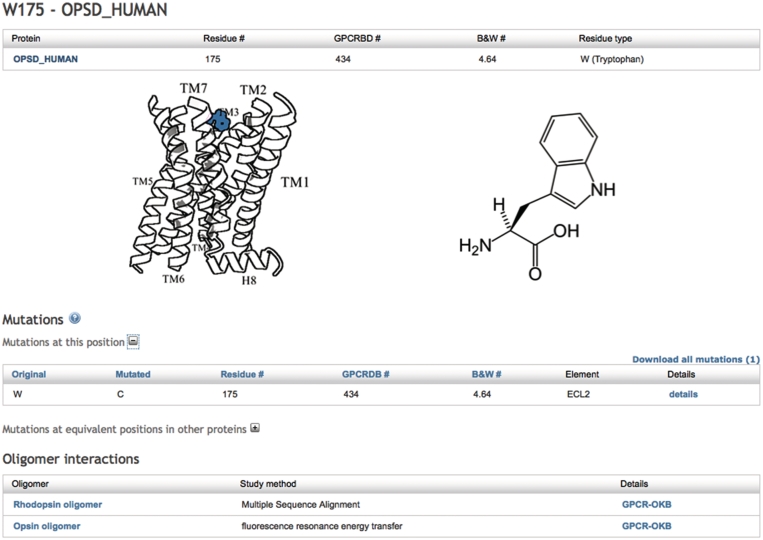
Screenshot of the detail page of residue W175 in OPSD_HUMAN. Residue numbers in different formats are shown, the approximate location of the residue is visually indicated and available mutations and oligomer data for this residue are listed.
Mutations can be found via the mutant search page, where one can search by residue number (multiple numbering schemes are available) and/or residue types. The GPCRDB offers a BLAST service that allows users to BLAST their sequence against the sequences in the GPCRDB.
All search options and the BLAST services are available via the web interface and as web services. A full SQL search facility will be made available in the near future to allow for complex queries and analyses.
Inferences
The amount of available GPCR related data is too large for a human to grasp and disseminate. The GPCRDB contains a series of inference engines that determine interesting correlations between the data, while a series of software tools help the user with data reduction and abstraction.
Building alignments
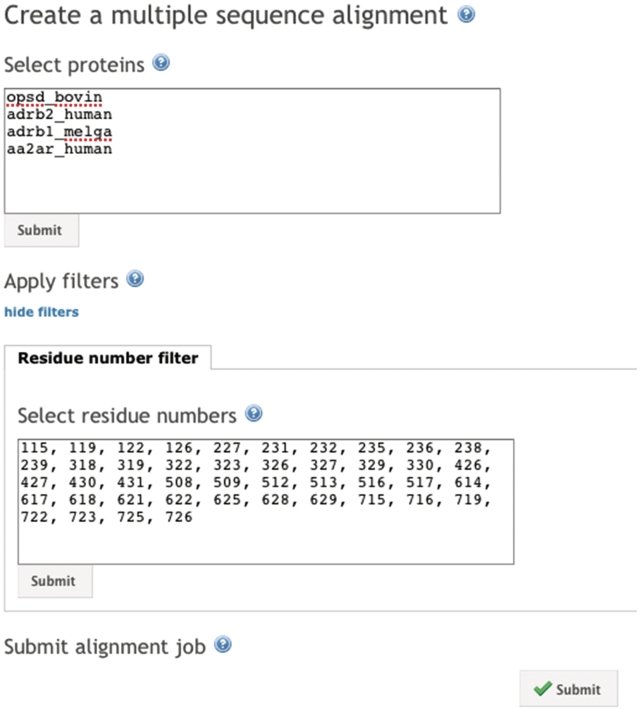
The GPCRDB offers the possibility to create custom-made alignments. The alignments are created by using the procedure that is used for the parent GPCR families as discussed earlier. Users can select the proteins and residue positions that should be aligned, allowing for the creation of e.g. an alignment of all binding pocket residues for a selection of proteins (Figure 7). The custom-built alignments are available for download and users can directly interact with the alignments using JalView.
Figure 6.
The protein search page.
Predicting the effects of mutations
We have started to create a service where users can predict the effects of a point mutation. As for now, predictions are mainly based on human knowledge that is stored in a computer readable format. This information is combined with a number of simple analyses on a homology model of the receptor being mutated, such as looking for steric clashes and helix disruptions. Results are presented as text-fragments that explain the effects of the mutation on the structure. Care has been taken to ensure that the results are presented in a life scientist friendly manner. The text contains references to literature and is enriched with figures and animations of the mutation and its surrounding environment. In the near future more intelligence will be added to the software, such as incorporating the quantitative data from the mutations extracted from literature and ligand-binding information.
Analysis of entropy derived patterns
We offer a page where users can interactively analyze the protein family alignments (Figure 2). Plots displaying entropy and variability scores are displayed with a 3D model and a MSA. Residues are linked in the three page elements, so that clicking on a residue position in the multiple sequence alignment will highlight the residue position in the structure as well as in the entropy and variability plots. This offers researchers a very intuitive way of looking at conservation scores, even at the subfamily or receptor level, and relating those scores to the 3D structure. In combination with above mentioned accessible data site directed mutagenesis candidate selection, homology modeling and ligand-binding hypotheses generation can be performed.
Annotating scientific literature
We have developed a new interface for the GPCR data in the form of a GPCR-specific PDF reader (manuscript in preparation). This reader can annotate scientific literature on GPCRs on the fly, providing users with context sensitive data from the GPCRDB (Figure 8). This software is available upon request and will be made freely available at the day of publication of this article.
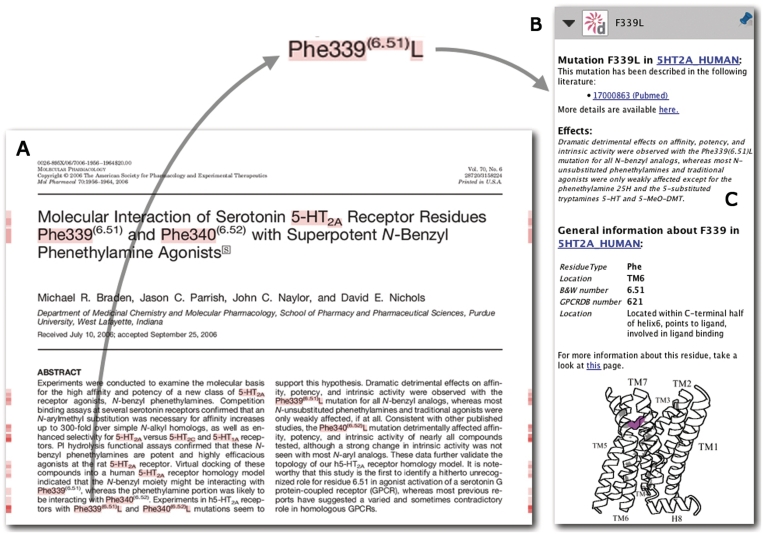
Figure 8.
An impression of the PDF reader [Utopia Documents (43), Utopia Documents-GPCRDB (in preparation)] interface to the GPCRDB data. On the left side (A) a scientific paper (44) is shown that is annotated by the GPCRDB. Annotations are available for all the highlighted words. On the right side (B) an example of such an annotation (the mutation F339L) is displayed. A short, manually extracted description of the effects of this mutation is included (C).
Figure 7.
A list of proteins and an optional list of residue positions can be used to generate custom alignments. In this figure we have selected a number of proteins for which crystal structures are available. The GPCR-binding pocket residue positions as proposed by Gloriam et al. (42) are used. The result will be an alignment of all pocket residues of the selected proteins.
IMPLEMENTATION
The data in the GPCRDB is stored in a PostgreSQL (http://www.postgresql.org/) relational database. The web service interface is developed with the Apache CXF (http://cxf.apache.org/) web services framework. We offer both SOAP and REST endpoints. The web interface is built using the Apache Wicket (http://wicket.apache.org/) web application framework. The database is accessed via a Hibernate (http://www.hibernate.org) object-relational mapping layer. The server is running within Sun’s Glassfish (http://glassfish.org) application server.
FUTURE DIRECTIONS
In the near future we would like to extend the interactive facilities of the GPCRDB by offering users more tools to analyze the available data. The entropy-variability analysis pages are a good example of the types of services we will be offering. In addition to our main focus of data collection and integration we would like to extend our focus towards the more challenging field of knowledge integration. The mutation effect predictor is a pilot project to explore the things we can do by combining human expertise with computational power. We are in the process of transforming the GPCRDB from mainly a one-stop resource for GPCRDB data to a place where scientists can use tools to interact with the data and make predictions.
FUNDING
Funding for open access charge: BioRange program of these Netherlands Bioinformatics Centre (NBIC); Netherlands Genomics Initiative (NGI), BSIK grant; European Commission within its FP6 Programme (to EMBRACE); under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LHSG-CT-2004-512092; This work was performed within the framework of the Dutch Top Institute Pharma project D1-105, The GPCR forum.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Bob Bywater for stimulating discussions, and Maarten Hekkelman and Tim te Beek for their support with computer science issues. Steve Pettifer, Dave Thorne and Phil McDermott are thanked for providing us with the Utopia Documents PDF reader and their support with connecting it to the GPCRDB.
REFERENCES
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, et al. The G protein-coupled receptor repertoires of human and mouse. Proc. Natl Acad. Sci. USA. 2003;100:4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Milligan G. G protein-coupled receptor hetero-dimerization: contribution to pharmacology and function. Br. J. Pharmacol. 2009;158:5–14. doi: 10.1111/j.1476-5381.2009.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoesser G, Baker W, van den Broek A, Camon E, Garcia-Pastor M, Kanz C, Kulikova T, Lombard V, Lopez R, Parkinson H, et al. The EMBL nucleotide sequence database. Nucleic Acids Res. 2001;29:17–21. doi: 10.1093/nar/29.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kourist R, Jochens H, Bartsch S, Kuipers R, Padhi SK, Gall M, Böttcher D, Joosten H, Bornscheuer UT. The alpha/beta-hydrolase fold 3DM database (ABHDB) as a tool for protein engineering. Chembiochem. 2010;11:1635–1643. doi: 10.1002/cbic.201000213. [DOI] [PubMed] [Google Scholar]
- 11.Kuipers RK, Joosten H, van Berkel WJH, Leferink NGH, Rooijen E, Ittmann E, van Zimmeren F, Jochens H, Bornscheuer U, Vriend G, et al. 3DM: systematic analysis of heterogeneous superfamily data to discover protein functionalities. Proteins. 2010;78:2101–2113. doi: 10.1002/prot.22725. [DOI] [PubMed] [Google Scholar]
- 12.Horn F, Bettler E, Oliveira L, Campagne F, Cohen FE, Vriend G. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hekkelman ML, Vriend G. MRS: a fast and compact retrieval system for biological data. Nucleic Acids Res. 2005;33:W766–W769. doi: 10.1093/nar/gki422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornton J. Annotations for all by all - the BioSapiens network. Genome Biol. 2009;10:401. doi: 10.1186/gb-2009-10-2-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez RC, Quinn AF, Garcia A, Labarga A, O’Neill K, Martinez F, Salazar GA, Hermjakob H. Dasty2, an Ajax protein DAS client. Bioinformatics. 2008;24:2119–2121. doi: 10.1093/bioinformatics/btn387. [DOI] [PubMed] [Google Scholar]
- 18.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute's data resources. Nucleic Acids Res. 2010;38:D17–D25. doi: 10.1093/nar/gkp986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. GLIDA: GPCR–ligand database for chemical genomics drug discovery–database and tools update. Nucleic Acids Res. 2008;36:D907–D912. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeman P. Drug dissociation constants for neuroreceptors and transporters. Receptor Tables. 1993;Vol 2 SZ Research, Toronto, Ontario. [Google Scholar]
- 21.Cutler D, Barbier A. In brief. Trends Pharmacol. Sci. 2002;23:258–259. [PubMed] [Google Scholar]
- 22.Edvardsen O, Reiersen AL, Beukers MW, Kristiansen K. tGRAP, the G-protein coupled receptors mutant database. Nucleic Acids Res. 2002;30:361–363. doi: 10.1093/nar/30.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horn F, Lau AL, Cohen FE. Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics. 2004;20:557–568. doi: 10.1093/bioinformatics/btg449. [DOI] [PubMed] [Google Scholar]
- 24.Joosten R, Salzemann J, Bloch V, Stockinger H, Berglund A, Blanchet C, Rudloff E, Combet C, Da Costa A, Deleage G, et al. {PDB_REDO: automated re-refinement of X-ray structure models in the PDB} J. App. Crystallogr. 2009;42:376–384. doi: 10.1107/S0021889809008784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohse MJ. Dimerization in GPCR mobility and signaling. Curr. Opin. Pharmacol. 2010;10:53–58. doi: 10.1016/j.coph.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Dean MK, Higgs C, Smith RE, Bywater RP, Snell CR, Scott PD, Upton GJ, Howe TJ, Reynolds CA. Dimerization of G-protein-coupled receptors. J. Med. Chem. 2001;44:4595–4614. doi: 10.1021/jm010290+. [DOI] [PubMed] [Google Scholar]
- 27.Hébert TE, Bouvier M. Structural and functional aspects of G protein-coupled receptor oligomerization. Biochem. Cell Biol. 1998;76:1–11. doi: 10.1139/bcb-76-1-1. [DOI] [PubMed] [Google Scholar]
- 28.Khelashvili G, Dorff K, Shan J, Camacho-Artacho M, Skrabanek L, Vroling B, Bouvier M, Devi LA, George SR, Javitch JA, et al. GPCR-OKB: the G protein coupled receptor oligomer knowledge base. Bioinformatics. 2010;26:1804–1805. doi: 10.1093/bioinformatics/btq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 30.Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Fouapproaches that performed well in CASP8. Proteins. 2009;77(Suppl. 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveira L, Paiva ACM, Vriend G. Correlated mutation analyses on very large sequence families. Chembiochem. 2002;3:1010–1017. doi: 10.1002/1439-7633(20021004)3:10<1010::AID-CBIC1010>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 32.Oliveira L, Paiva AC, Sander C, Vriend G. A common step for signal transduction in G protein-coupled receptors. Trends Pharmacol. Sci. 1994;15:170–172. doi: 10.1016/0165-6147(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 33.Oliveira L, Paiva PB, Paiva ACM, Vriend G. Sequence analysis reveals how G protein-coupled receptors transduce the signal to the G protein. Proteins. 2003;52:553–560. doi: 10.1002/prot.10489. [DOI] [PubMed] [Google Scholar]
- 34.Folkertsma S, van Noort P, Van Durme J, Joosten H, Bettler E, Fleuren W, Oliveira L, Horn F, de Vlieg J, Vriend G. A family-based approach reveals the function of residues in the nuclear receptor ligand-binding domain. J. Mol. Biol. 2004;341:321–335. doi: 10.1016/j.jmb.2004.05.075. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira L, Paiva PB, Paiva ACM, Vriend G. Identification of functionally conserved residues with the use of entropy-variability plots. Proteins. 2003;52:544–552. doi: 10.1002/prot.10490. [DOI] [PubMed] [Google Scholar]
- 36.Ye K, Lameijer EM, Beukers MW, Ijzerman AP. A two-entropies analysis to identify functional positions in the transmembrane region of class A G protein-coupled receptors. Proteins. 2006;63:1018–1030. doi: 10.1002/prot.20899. [DOI] [PubMed] [Google Scholar]
- 37.Oliveira L, Paiva A, Vriend G. A common motif in G-protein-coupled seven transmembrane helix receptors. J. Comp. Aided Mol. Des. 1993;7:649–658. [Google Scholar]
- 38.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goble CA, Bhagat J, Aleksejevs S, Cruickshank D, Michaelides D, Newman D, Borkum M, Bechhofer S, Roos M, Li P, et al. myExperiment: a repository and social network for the sharing of bioinformatics workflows. Nucleic Acids Res. 2010;38(Suppl.):W677–W682. doi: 10.1093/nar/gkq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, et al. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gloriam DE, Foord SM, Blaney FE, Garland SL. Definition of the G protein-coupled receptor transmembrane bundle-binding pocket and calculation of receptor similarities for drug design. J. Med. Chem. 2009;52:4429–4442. doi: 10.1021/jm900319e. [DOI] [PubMed] [Google Scholar]
- 43.Attwood TK, Kell DB, McDermott P, Marsh J, Pettifer SR, Thorne D. Utopia documents: linking scholarly literature with research data. Bioinformatics. 2010;26:i568–i574. doi: 10.1093/bioinformatics/btq383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braden MR, Parrish JC, Naylor JC, Nichols DE. Molecular interaction of serotonin 5-HT2A receptor residues Phe339(6.51) and Phe340(6.52) with superpotent N-benzyl phenethylamine agonists. Mol. Pharmacol. 2006;70:1956–1964. doi: 10.1124/mol.106.028720. [DOI] [PubMed] [Google Scholar]