Abstract
Fast viral adaptation and the implication of this rapid evolution in the emergence of several new infectious diseases have turned this issue into a major challenge for various research domains. Indeed, viruses are involved in the development of a wide range of pathologies and understanding how viruses and host cells interact in the context of adaptation remains an open question. In order to provide insights into the complex interactions between viruses and their host organisms and namely in the acquisition of novel functions through exchanges of genetic material, we developed the PhEVER database. This database aims at providing accurate evolutionary and phylogenetic information to analyse the nature of virus–virus and virus–host lateral gene transfers. PhEVER (http://pbil.univ-lyon1.fr/databases/phever) is a unique database of homologous families both (i) between sequences from different viruses and (ii) between viral sequences and sequences from cellular organisms. PhEVER integrates extensive data from up-to-date completely sequenced genomes (2426 non-redundant viral genomes, 1007 non-redundant prokaryotic genomes, 43 eukaryotic genomes ranging from plants to vertebrates) and offers a clustering of proteins into homologous families containing at least one viral sequences, as well as alignments and phylogenies for each of these families. Public access to PhEVER is available through its webpage and through all dedicated ACNUC retrieval systems.
INTRODUCTION
Viruses are responsible for a large number of infectious diseases and cancers. Recently, new viral diseases have emerged leading to severe consequences on human activities. The emergence of many of these new viruses can be attributed to recombining viruses as well as to host species jump (1–3). Therefore, understanding how viruses interact with their hosts and more specifically how the complex interactions between viruses and their host organisms are acquired and maintained throughout evolution, remains a major challenge (4–7). In order to assess this question, it is of prime importance to be able to detect and quantify the occurrence of lateral gene transfer events, and the impact of these events on viral–host co-evolution. Indeed, the mechanisms behind fast viral adaptation are far from being elucidated. Thus, we developed a global approach aimed at providing accurate evolutionary and phylogenetic information to tackle these questions.
The major drawback of currently available databases of homologous families to the study of viral homologies and lateral gene transfer in viruses is their taxonomic compartimentalization. Indeed, current databases present families of homologies either restricted to viruses only [Protein Clusters (8), GeneTree (9)] or to viral taxonomic groups [Viral Orthologous Cluster (10)], some also not presenting viral information [HomoloGene (11)]. The few databases that do present viral and non-viral sequences, such as Pfam (12) or the Conserved Domain Database (13) do not provide complete phylogenetic trees. This translates into the fact that it is not currently possible to have a global view on viral–host lateral gene transfers due to the difficulty of obtaining global information on cross-taxa transfers at the viral level.
We present the first public release of PhEVER, a unique database of homologous gene families containing sequences (i) of all completely sequenced viruses and (ii) from fully sequenced cellular organisms. The protein sequences are clustered—without a priori and according to similarity criteria—into families containing either only viral sequences or both viral and cellular sequences. PhEVER integrates extensive data from up-to-date completely sequenced genomes spanning a wide taxonomic range (2426 non-redundant viral genomes, 1007 non-redundant prokaryotic genomes, 43 eukaryotic genomes ranging from plants to vertebrates). To our knowledge, this is the most complete database of families of homologous viral sequences. Indeed, it not only spans all known viral groups but it also has the unique feature of presenting homologies with eukaryotic and prokaryotic sequences. The database offers a clustering of proteins into homologous families containing at least one viral sequence, as well as pre-computed alignments and phylogenies for each of these families. Alignments and phylogenies are built according to state-of-the-art phylogeny procedures and we provide tools to edit them and recompute them on the fly (14). We also provide the possibility for users to assign their sequence of interest to a family and to re-build the phylogeny accordingly through the HoSeqI tool (15). PhEVER thus constitutes a comprehensive working tool to detect sequence homologies and possible gene transfer events. Public access and documentation is available through the database webpage and through all dedicated ACNUC retrieval systems (16).
FAMILIES OF VIRAL PROTEINS
We developed a genome-wide cross-taxa approach to build a database of families of homologous sequences and to provide accurate alignments and phylogenies for the constructed families. The layout of the implementation of the PhEVER database is represented in Figure 1 and the details concerning its sequence content are available in Table 1. In order to avoid redundancy due to the availability of numerous genomes of similar bacterial and viral strains in public databases, we collected all completely sequenced viral and bacterial genomes from RefSeq Viral (17) and Genome Reviews (18), two non-redundant and curated databases of completely sequenced genomes. To this high-quality curated data composed of Archaea, Bacteria and Eukarya, we added nine eukaryotic genomes from Ensembl (Aedes aegypti, Anopheles gambiae, Bos taurus, Caenorhabditis elegans, Danio rerio, Drosophila melanogaster, Gallus gallus, Homo sapiens, Mus musculus) (19) to allow for a large representation of species from the different domains of life. The PhEVER database was structured under the ACNUC system (20) allowing it to be queried using a web interface and a large number of tools specifically developed for this database management system (14).
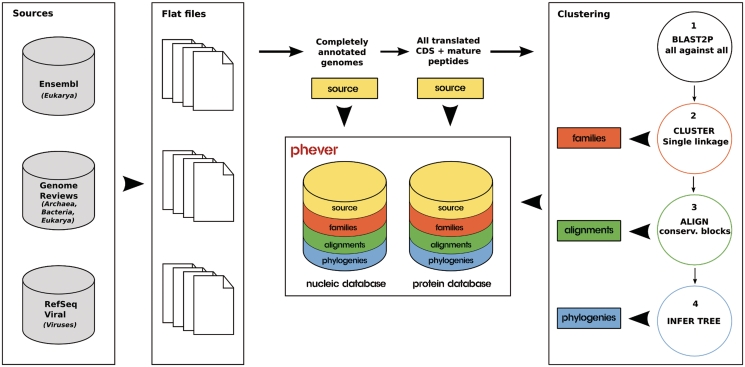
Figure 1.
Flow chart of the PhEVER building process. Complete genomes and their annotations were retrieved from three external public databases (Ensembl, Genome Reviews and RefSeq Viral) to provide high-quality non-redundant data for Eukarya, Archaea, Bacteria and Viruses. Two databases (nucleic acids, proteins) were constructed from this data to form PhEVER. All annotated CDS and mature peptides were translated and used for the clustering procedure. The homologous families thus produced were annotated in PhEVER, and alignments and phylogenies were built for each family and incorporated in the databases.
Table 1.
Global species and sequence content in PhEVER
| No. of species | No. of proteinsa | No. of proteins in fam.b | Data source | |
|---|---|---|---|---|
| All | 3476 | 4 515 271 | 333 618 (1%) | – |
| Viruses | 2426 | 82 929 (2718 m.p.c) | 82 784 (100%) | RefSeq |
| Bacteria | 937 | 3 207 914 | 232 066 (7%) | Genome Reviews |
| Archaea | 70 | 158 919 | 6702 (4%) | Genome Reviews |
| Eukarya | 43 | 1 065 509 | 12 066 (1%) | Ensembl + Genome Reviews |
| Eukarya (excl. Anopheles)d | – | 580 567 | 11 201 (2%) | – |
aProteins correspond to translated annotated CDS. The number of proteins in the protein database is therefore equal to the number of CDS in the nucleic database.
bNumber of proteins associated to a family, followed by the proportion of proteins associated to a family in the taxonomic group.
c2718 mature peptides are added to the 80 210 proteins translated from all CDS.
dThe genome of A. gambiae contains data from different haplotypes and presents therefore a high level of redundancy.
From the flat files containing the genomes and their annotations, two databases were built under the ACNUC database management system (20), which is specifically aimed at building, storing and querying biological sequence data. One of them contains the nucleic sequences, the other contains the proteins generated by translating all CDS of the complete genomes—using the appropriate genetic codes. For viral genomes presenting polypeptides which further maturate in vivo into mature peptides, the mature peptides were added to the set of translated proteins according to the annotations specified in the given genome (Figure 1). Table 1 lists the global content of the databases as well as their original sources. Annotations were extracted from UniProtKB via the cross-references found in the CDS (21). Subsequently, sequences in the nucleic and protein PhEVER database were clustered into families and were assigned a family accession number. Alignments and phylogenies were built for each of these families according to state-of-the-art procedures (Figure 1). The classification of all organisms present in the database was retrieved from the taxonomy database at National Centre for Biotechnology Information (11) and is available on the PhEVER web interface.
CLUSTERING INTO FAMILIES
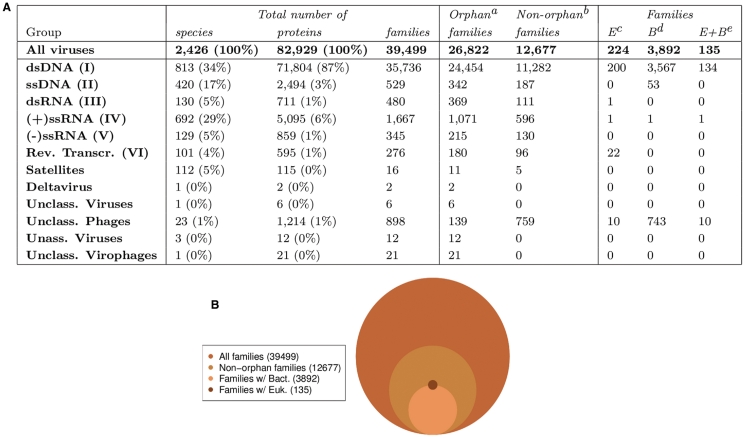
The clustering of proteins into homologous families was constructed using an automated procedure similar to the one described in (14) and implemented within a parallel framework in a software package called SiLiX. Briefly, sequences were assigned to protein families by simple transitive link using the following criteria. A similarity search of all translated proteins and mature peptides against all was performed using BLASTP2 similarity search with the BLOSUM62 substitution matrix, a 10−4 e-value threshold and the ‘m S’ filter option (22,23). For each pair of sequences, HSPs which were not compatible with a global alignment were removed. Two sequences were included in the same family if the sum of the remaining HSPs covered >80% of the proteins length (and at least 100 amino acids) and if their identity was ≥35%. These two criteria were previously shown to provide a good trade-off between the ability of clustering sequences from divergent organisms and the quality of resulting alignments for subsequent phylogenetic analyses (14). Finally, only families containing at least one viral sequence were integrated in the database. For each of the families, a small description built from the gene annotations ordered by frequency is available on the web interface. Figure 2 presents the distribution of viral species, proteins and families according to each viral group (A) as well as a Venn diagram representing the families content (B). Figure 2A presents PhEVER’s broad taxonomical distribution covering all Baltimore groups (24). This distribution is naturally biased towards dsDNA, ssDNA and positive-sense ssRNA viruses reflecting the bias in genomic sequencing efforts. Indeed these groups contain long-studied viral families—either for their medical interest or for their economical impacts—such as Caudovirales, Poxviridae, Herpesvirales, Flaviviridae, Picornavirales or Parvoviridae. Figure 2 (B) shows a large number of orphan families indicating that a significant proportion of viral proteins (32%) do not contain any homologs with proteins from known genomes. These proteins, among which some might possibly be caused by annotation errors, are unfit for comparative functional analysis and should be the focus of future experimental studies to validate them and to provide with crucial information on viral mechanisms. Figure 2B also shows the small number of families sharing sequences from both viruses and eukaryotes compared to the relatively high number of families sharing sequences from both viruses and bacteria. This observation may be due to different underlying biological mechanisms but might also be an indicator of a still low coverage of the Eukarya domain.
Figure 2.
Global information on PhEVER content. (A) Number of species, proteins, families in each viral group [groups correspond to the NCBI Taxonomy following the ICTV nomenclature (36); groups corresponding to the Baltimore classification are additionally indicated in Roman notation (24)]. (B) Venn diagram of the number of families, non-orphan families and families containing Bacteria and Eukarya. aOrphan families are families containing only one viral sequence and no other sequence. This sequence presents no sufficient similarity with any sequence of the database, bNon-orphan families are all families containing at least two sequences, cFamilies containing at least one viral sequence and one eukaryotic (E) sequence, dFamilies containing at least one viral sequence and one bacterial (B) sequence and eFamilies containing at least one viral sequence, one eukaryotic sequence and one bacterial sequence (E+B).
DETECTION OF HORIZONTAL GENE TRANSFER
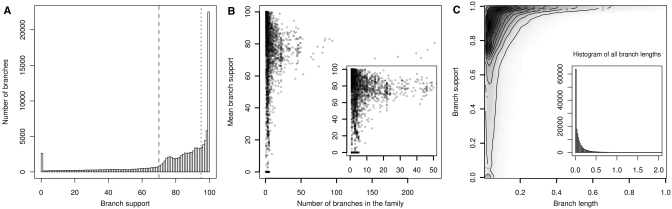
One of the applications of PhEVER is the detection of horizontal gene transfer events by comparing a gene family tree with the expected species tree. The discrepancies between the gene family history and the species phylogeny can then be an indication of possible events including a gene duplication, a gene loss or a horizontal gene transfer. The quality of the gene phylogenies is therefore essential and in this perspective, we implemented a procedure based on a rigorous methodology. First, the clustering into families was built with criteria leading to conservative families. Second, maximum likelihood phylogenetic trees were inferred for all families based on conserved aligned blocks. In our databases, for all families containing at least three sequences, pre-computed alignments and phylogenies are therefore already available. This allows for a simple and accurate overview of any family without the need of heavy computations and can be a useful tool to search for lateral gene transfers in viral genes. For each family containing at least three sequences, alignments were estimated using MUSCLE with default parameters (25). All alignments were treated with Gblocks (26) to select conserved blocks. Phylogenetic trees were inferred by maximum likelihood using PhyML (27) with a JTT evolutionary model (28). Branch support was inferred using the Shimodaira–Hasegawa-like non-parametric procedure implemented in PhyML (27,29). To accommodate for weak phylogenetic signal, a thorough exploration of the tree space was made through topological rearrangements using the Nearest Neighbor Interchange topology search method. Finally, for visualization purposes, the trees were then rooted using midpoint rooting. This procedure allowed us to build accurate phylogenies sustained by branches of high support values. Indeed, Figure 3 shows that ∼80% of all branch supports have a value higher than 75 and one-third are above 95. Global statistics are biased by the presence of low branch supports. These are mostly due to few small families of less than 10 leaves, indicating that most families present robust phylogenies (Figure 3B). Finally, Figure 3C indicates that the low branch support values are attributable to very similar sequences with small branch lengths and might be linked to unresolved topologies. By contrast, all branch lengths longer than 0.15 subst/site display a high branch support which reveals the accuracy of our phylogenetic inference procedure.
Figure 3.
Branch support in PhEVER trees. (A) Histogram of node supports in all phylogenetic trees. The dashed line indicates the support value of 70 (80% of the distribution is above this threshold), the dotted line indicates the support value of 95 (one-third of the distribution is above this threshold). (B) Relation between the mean node support in a family and the size of this family, measured by the number of nodes in the family. The inner figure shows the mean node support of the smaller families. (C) Relation between branch support and branch length. Only branch lengths inferior to 1 are plotted on this figure. The inner figure shows the distribution of branch lengths justifying the focus on branch lengths inferior to 1. For all figures, only trees with more than four leaves are presented here. Branches with length smaller than 10−5 were considered unresolved multifurcations and were discarded.
QUERY THE DATABASE
PhEVER is structured under the ACNUC sequence database management system and a large number of tools have been developed around this database management system [see (16) for an overview]. PhEVER can therefore be queried using (i) web applications, (ii) standalone software (iii) or embedded within Python, R or C code. The PhEVER web interface is available at http://pbil.univ-lyon1.fr/databases/phever and allows to search for sequences or families by combining several criteria (including species, gene names, annotation terms) as well as by crossing taxa. The graphical user interface QUERY_WIN and the terminal-based interface Raa_query (16) implement more features than the web interface and allow for remote ACNUC access and query as well as for the automatization of querying processes through standalone software. Finally the C language API, Python language API (16) and the seqinR package for R (30) implement tools for integrating queries in user designed code. Note that the PhEVER database can also be installed on a local machine or server for fastest data access. All files necessary for the PhEVER installation are provided through our ftp website or by simple request.
PHEVER WEB INTERFACE
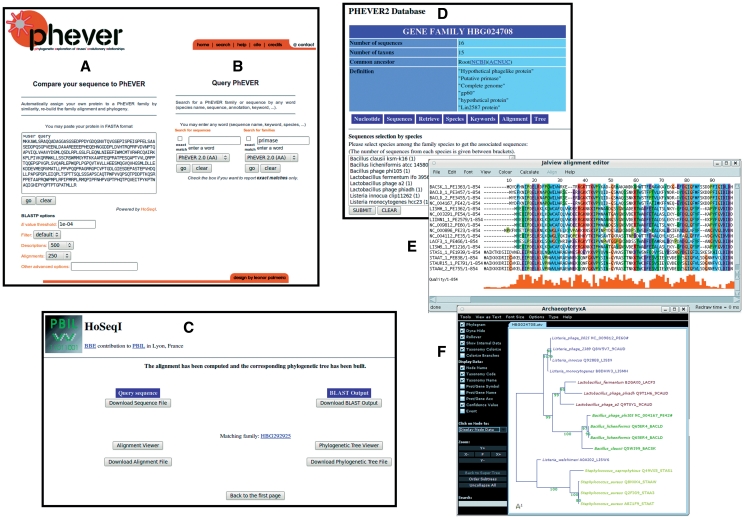
The PhEVER web interface allows for two query forms represented in Figure 4A and B. On the one hand (Figure 4A), the HoSeqI tool allows to search in PhEVER families with a user provided query (15). This query is used to BLAST the proteins present in PhEVER and to match the most related family. Alignments and phylogenies for this new family containing the user provided sequence can be recomputed on the fly, visualized and manipulated with Java applets (Figure 4F) (31,32). On the other hand (Figure 4B), the query tool allows to directly query the database with a very diverse range of terms including gene name, annotation term, species name, protein accession number, genome accession number and family accession number. The species and sequences represented in each family detected by the query can then be viewed (Figure 4D) as well as the alignments and phylogenies which can be edited online via Java applets (Figure 4E) (31,32). More details on how to query PhEVER are presented in the Supplementary Data.
Figure 4.
Overview of some of the possibilities of the PhEVER interface. (A) HoSeqI tool to query PhEVER families with a user provided sequence, (B) Query the database with terms including gene name, species name, annotation terms, (C) Alignments and phylogenies can be recomputed on the fly through the HoSeqI tool, (D) Visualization of a PhEVER family and the species and sequences composing it, (E) JalView applet to edit the pre-computed alignments and (F) ATV applet to edit the pre-computed phylogenies.
CONCLUSION AND PERSPECTIVES
PhEVER is the first open access database to provide information at the cross-taxa scale for the analysis of virus–virus and virus–host protein transfers. It compiles information from all kingdoms of life, and handles data from the genomes of all completely sequenced viruses and prokaryotes and of a large range of eukaryotes. It is the largest database of viral homologous families and offers highly accurate pre-computed alignments and phylogenies, making it a powerful tool for the analysis of horizontal gene transfer and more widely for the analysis of gene history.
Our objective is to continue the development of PhEVER around the analysis of protein evolution in the context of virus–virus and virus–host interactions. More specifically, the next step we have under development is the detection of evolutionary conserved modules in the proteins present in PhEVER. Indeed, there is strong evidence that proteins evolve in a modular way, where modules are defined as parts of proteins sharing a common evolutionary history. These modules act as small interchangeable blocks of sequences that may be combined into proteins and form novel functions (33–35). We are interested in providing a global tool allowing to analyse the weight of modular evolution in viral adaptation. We will therefore implement the detection of modules in PhEVER proteins to provide information concerning the exchanges of genetic information at the sub-protein level.
In conclusion, PhEVER aims at being a comprehensive tool for the analysis of virus–virus and virus–host relationships from an evolutionary point of view, namely through the analysis of genomic interchanges. It should become a valuable tool for anyone working on viral evolution, but also to understand the general mechanisms behind protein evolution and functional innovation.
DATABASE ACCESS AND UPDATES
Public access and documentation is freely available through the database webpage (http://pbil.univ-lyon1.fr/databases/phever/) and through all dedicated ACNUC retrieval systems. More information on dedicated ACNUC retrieval systems, such as standalone query software (16), the seqinR package for R (30) or the C and Python APIs, can be obtained in the Supplementary Data or on the PhEVER webpage. The PhEVER flat files for local installation are available through our ftp server (ftp://pbil.univlyon1.fr/pub/phever) and instructions are available on the database webpage. The PhEVER database is updated every 6 months. This update frequency allows to follow the fast pace of viral and prokaryotic genome sequencing as well as to obtain updated genomic annotations for large eukaryotic genomes. Previous versions of the database remain available upon request.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Interaubio project, granted by the Région Rhône-Alpes. The Région Rhône-Alpes and the University of Liège (to L.P.). Funding for open access charge: BAMBOO Team, INRIA Rhône-Alpes.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Vincent Navratil and Vincent Daubin for helpful dicussions as well as Linda Artmann for assisting the figure preparation. The authors also acknowledge the CC IN2P3 (Villeurbanne) for the computing resources and Pascal Calvat for his technical help as well as the computer department at PBIL-DOUA and LBBE for assistance and maintenance of the PhEVER server.
REFERENCES
- 1.Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO, Hahn BH. The origins of acquired immune deficiency syndrome viruses: where and when? Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 2001;356:867–876. doi: 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes EC. The comparative genomics of viral emergence. Proc. Natl Acad. Sci. USA. 2010;107(Suppl. 1):1742–1746. doi: 10.1073/pnas.0906193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shackelton LA, Holmes EC. The evolution of large DNA viruses: combining genomic information of viruses and their hosts. Trends Microbiol. 2004;12:458–465. doi: 10.1016/j.tim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Curr. Opin. Microbiol. 2005;8:444–450. doi: 10.1016/j.mib.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Filée J. Lateral gene transfer, lineage-specific gene expansion and the evolution of Nucleo Cytoplasmic Large DNA viruses. J. Invertebr. Pathol. 2009;101:169–171. doi: 10.1016/j.jip.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Hughes AL, Irausquin S, Friedman R. The evolutionary biology of poxviruses. Infect. Genet. Evol. 2010;10:50–59. doi: 10.1016/j.meegid.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O’Neill K, Resch W, Resenchuk S, et al. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 2009;37(Database issue):D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Y, Dickerman AW. GeneTrees: a phylogenomics resource for prokaryotes. Nucleic Acids Res. 2007;35(Database issue):D328–D331. doi: 10.1093/nar/gkl905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehlers A, Osborne J, Slack S, Roper RL, Upton C. Poxvirus Orthologous Clusters (POCs) Bioinformatics. 2002;18:1544–1545. doi: 10.1093/bioinformatics/18.11.1544. [DOI] [PubMed] [Google Scholar]
- 11.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38(Database issue):D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penel S, Arigon A-M, Dufayard J-F, Sertier A-S, Daubin V, Duret L, Gouy M, Perrière G. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009;10(Suppl. 6):S3. doi: 10.1186/1471-2105-10-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arigon A-M, Perrière G, Gouy M. HoSeqI: automated homologous sequence identification in gene family databases. Bioinformatics. 2006;22:1786–1787. doi: 10.1093/bioinformatics/btl179. [DOI] [PubMed] [Google Scholar]
- 16.Gouy M, Delmotte S. Remote access to ACNUC nucleotide and protein sequence databases at PBIL. Biochimie. 2008;90:555–562. doi: 10.1016/j.biochi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37(Database issue):D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterk P, Kulikova T, Kersey P, Apweiler R. The EMBL Nucleotide Sequence and Genome Reviews Databases. Methods Mol. Biol. 2007;406:1–21. doi: 10.1007/978-1-59745-535-0_1. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouy M, Gautier C, Attimonelli M, Lanave C, di Paola G. ACNUC–a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. CABIOS. 1985;1:167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- 21.UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37(Database issue):D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Wootton J, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 24.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 27.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 28.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 29.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- 30.Charif D, Lobry J. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural Approaches to Sequence Evolution: Molecules, Networks, Populations. NY: Springer; 2007. pp. 207pp. 207–232. [Google Scholar]
- 31.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 32.Zmasek CM, Eddy SR. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 33.Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 34.Moore AD, Björklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 2008;33:444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Trifonov EN, Frenkel ZM. Evolution of protein modularity. Curr. Opin. Struct. Biol. 2009;19:335–340. doi: 10.1016/j.sbi.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Fauquet C, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. Oxford, UK: Elsevier Academic Press; 2005. [Google Scholar]