Abstract
Reactome (http://www.reactome.org) is a collaboration among groups at the Ontario Institute for Cancer Research, Cold Spring Harbor Laboratory, New York University School of Medicine and The European Bioinformatics Institute, to develop an open source curated bioinformatics database of human pathways and reactions. Recently, we developed a new web site with improved tools for pathway browsing and data analysis. The Pathway Browser is an Systems Biology Graphical Notation (SBGN)-based visualization system that supports zooming, scrolling and event highlighting. It exploits PSIQUIC web services to overlay our curated pathways with molecular interaction data from the Reactome Functional Interaction Network and external interaction databases such as IntAct, BioGRID, ChEMBL, iRefIndex, MINT and STRING. Our Pathway and Expression Analysis tools enable ID mapping, pathway assignment and overrepresentation analysis of user-supplied data sets. To support pathway annotation and analysis in other species, we continue to make orthology-based inferences of pathways in non-human species, applying Ensembl Compara to identify orthologs of curated human proteins in each of 20 other species. The resulting inferred pathway sets can be browsed and analyzed with our Species Comparison tool. Collaborations are also underway to create manually curated data sets on the Reactome framework for chicken, Drosophila and rice.
INTRODUCTION
Reactome is an open source, open access, manually curated, peer-reviewed pathway database of human pathways and processes (1). Pathway annotations are created by expert biologists, in collaboration with Reactome editorial staff and cross-referenced to proteins (UniProt) and genes (NCBI EntrezGene, Ensembl, UCSC and HapMap), small molecules (KEGG Compound and ChEBI), primary research literature (PubMed) and GO controlled vocabularies (2–9). The Reactome data model generalizes the concept of a reaction to include transformations of entities such as transport from one compartment to another and interaction to form a complex, as well as the chemical transformations of classical biochemistry. Entities include nucleic acids, small molecules, proteins (with or without post-translational modifications) and macromolecular complexes. This generalization permits the capture of a range of biological processes that spans signaling, metabolism, transcriptional regulation, apoptosis and synaptic transmission in a single internally consistent, computationally navigable format. Reactome is an all-inclusive resource of human pathways for basic research, genome analysis, pathway modeling, systems biology and education. In the past 2 years, the Reactome data set has nearly doubled in size and new tools for data aggregation and data analysis have become available. To support the continued development of the Reactome knowledgebase, we have redesigned the Reactome web site and data analysis software.
Expanded pathway coverage
Reactome’s recruitment of expert authors and curators has given us access to key aspects of human biology. The current release of Reactome (Version 34, September 2010) describes the roles of 5272 human proteins (26% of the 20 286 human SwissProt entries) and 3504 macromolecular complexes in 3847 reactions organized into 1057 pathways. Over the last year, we have added new higher order topics. Notable additions include the molecular anatomy of transcriptional regulation, a largely complete catalogue of receptors with known ligands involved in GPCR signaling (10), Toll-like Receptors, Chromosome maintenance, Olfactory Signaling, Myogenesis, N-Glycan biosynthesis and Metabolism of RNA. Reactome has prototyped additional types of annotations to support pathway curation. We have curated pathways relating to insulin signaling cycle to prototype pathway–disease annotations. We have also developed a framework for physiological process annotations, e.g. vesicle transport and glutamate-mediated neurotransmission. To support the creation of the new pathway diagrams, we defined canonical pathways corresponding to 161 discrete biological domains. This enabled a simplification of the event hierarchy in the new Pathway Browser and has minimized event sharing across different pathways. Visualization features were implemented in the Author and Curator Tool to enable the layout and editing of these new pathways diagrams as part of our curation process.
The Reactome data model has been extended to the manual curation of pathways in model organisms. Gallus Reactome (http://gallus.reactome.org), an effort led by Carl Schmidt of the University of Delaware has been modeled after Reactome. The first public release was in mid-2009 and now Gallus Reactome includes annotations for 127 reactions involving 133 Gallus proteins in the domains of intermediary metabolism and DNA repair. A collaboration among Reactome, Michael Ashburner and Mark Williams at Cambridge University, similarly uses Reactome software and hardware to create and maintain a Drosophila pathway database (http://fly.reactome.org/). Its third release went public in mid-2010, and includes data for Wingless, JAK/STAT, Imd, Toll, Hedgehog, Circadian Clock, Hippo/Warts and Planar Cell Polarity signaling pathways.
Redesigned reactome web site
The rapid growth in content has made the ‘starry sky’ reaction map display unwieldy as a navigation and visualization tool. At the same time, our outreach has grown to encompass diverse user groups interested in browsing a particular process or protein as a textbook, analyzing high-throughput expression data sets, data mining and data aggregation and online resources for education. Our front page has thus been redesigned to support quick, intuitive access to our data and tools as both features of the knowledgebase continue to grow. The new web site retains a comprehensive top menu bar that provides access to all of our tools and resources. It is now accompanied by a sidebar that provides access to basic, widely used tools for pathway browsing and data analysis, and panels that give a thumbnail overview of Reactome information, tutorials, recent news and a view of a recently added pathway of topical interest.
Improved pathway visualization
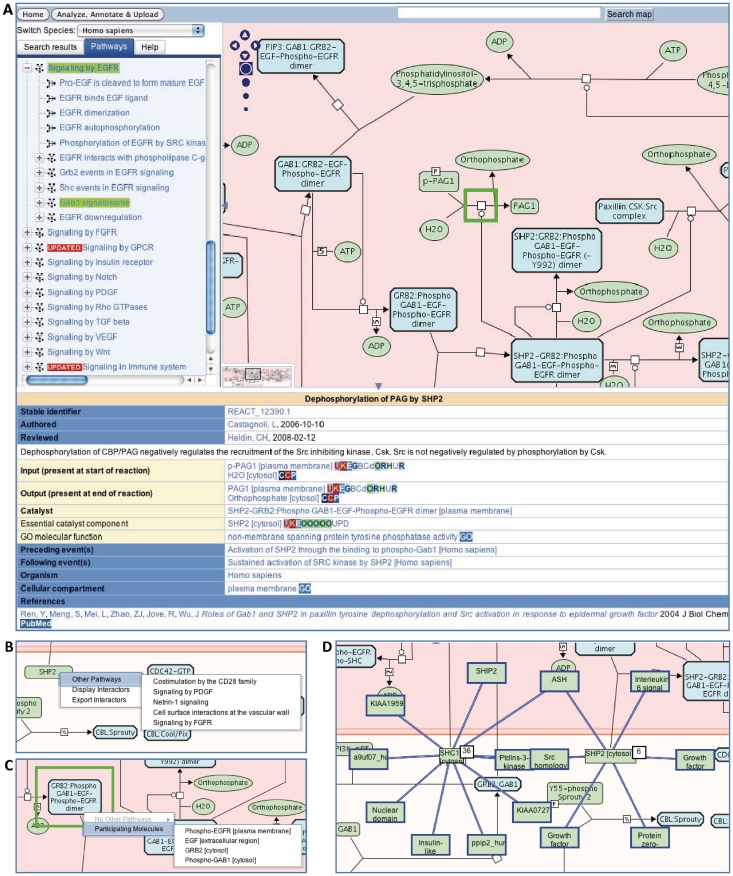
Visualization of full pathway information in a consistent format is vital to support the pathway-based analysis of complex experimental and computational data sets. To support such visual navigation and analysis of Reactome data we have developed, in collaboration with the ENFIN project (11), a new Pathway Browser based upon the Systems Biology Graphical Notation (SBGN) (12). SBGN is a standard graphical representation of biological pathway and network models, i.e. every molecule and reaction has a particular shape, color and cellular location. Our entire content has been organized into 161 canonical pathways, each displayed in this format. The Reactome Pathway Browser consists of four key elements. First, the ‘Search’ bar at the top of the page queries the entire Reactome database. Second, the ‘Pathways’ panel provides a scrolling display of the Reactome canonical pathway hierarchy. Third, clicking on the pathway name displays the corresponding pathway diagram in the ‘Visualization’ panel on the right side. The ‘Visualization’ panel offers interactive and dynamic pathway diagrams permitting zooming, scrolling and highlighting of events and molecules. Fourth, clicking on events and molecules in the pathway diagram uncovers a ‘Details’ panel below the pathway diagram with additional textual information about the events and molecules, respectively. Further functionality is provided in the form of context sensitive menus within the ‘Visualization’ panel (Figure 1). The precise features of the context sensitive menus are determined by the nature of the physical entity (small molecule, protein, complex): (i) a list of the other pathways in Reactome in which the selected entity participates; (ii) a display of the physical entities that contribute to the macromolecular complex; and, optionally (iii) a list of interactors of the entity from selected interaction databases (described later). The pathway diagrams are available for download as static PNG and PDF files. Dynamic pathway images compatible with third-party tools like Cytoscape (13) and CellDesigner (14) are currently being developed.
Figure 1.
The Reactome pathway browser and the molecular overlays. (A) The main features of the pathway browser are the ‘Search’ bar at the top, the ‘Pathways’ panel on the left, the ‘Visualization’ panel on the right and a ‘Details’ panel (hidden by default when the pathway browser first loads). When a pathway is selected from the ‘Pathways’ panel hierarchy, it is highlighted in bright green in the hierarchy and its parent terms are highlighted in yellow–green. A description of any reaction or pathway in the pathway diagrams can be displayed below the diagram in the ‘Details’ panel by selecting the event within the diagram. Right-clicking the mouse while the cursor is over a physical entity displays the context sensitive menus. (B) Selecting ‘Other Pathways’ displays additional Reactome pathways that contain the highlighted physical entity. Navigating to the additional pathway is achieved by clicking the pathway name. (C) Selecting ‘Participating Molecules’ displays additional components of the macromolecular complex. Displaying the component information in the ‘Details’ panel is achieved by clicking the entity name. (D) Protein–protein interactions are displayed in the pathway browser for SHC1 [cytosol] and SHP2 [cytosol] physical entities.
Integrating molecular interactions and networks onto reactome pathway diagrams
The Reactome data sets are a highly reliable platform for pathway-based data analysis but suffer from a low coverage of human proteins. To increase protein coverage and associated protein annotations, we have integrated molecular interaction (MI) data and network information into the Reactome pathway diagrams (Figure 1). The MI overlay displays proteins interacting with the manually annotated protein components of a Reactome pathway. As mentioned previously, individual protein interactors can be displayed using the context-dependent menus in the pathway ‘Visualization’ workspace. It is also possible to overlay all the interactors for all the pathway proteins by means of the ‘Analyze, Annotate and Upload’ feature of the Pathway Browser. The network overlay tool employs a PSICQUIC interface to implement flexible import of binary MI data into Reactome pathway diagrams. PSICQUIC is already widely implemented by interaction databases, including BioGRID, ChEMBL, IntAct, iRefIndex, MINT and STRING (15–20). The nodes and edges of the network overlay are interactive, providing links to the physical entity and interaction databases, respectively. Two of additional interaction data sets are managed by the Reactome group, ‘Reactome’ and ‘Reactome-FIs’. The original ‘Reactome’ data set reflects MI data derived from Reactome reactions and complexes. A new ‘Reactome-FIs’ (functional interactions) data set unites interactions from Reactome and those derived from other pathway databases, including KEGG, BioCyc, Panther, The Cancer Cell Map (http://cancer.cellmap.org/) and PID (7,21–23) with pair-wise interactions gleaned from physical protein–protein interactions in human and model organisms, gene co-expression data, protein domain–domain interactions, protein interactions generated from text mining and GO annotations (24). The ‘Reactome-FIs’ network contains 209 988 functional interactions encompassing 10 956 proteins (excluding splice isoforms), reflecting 46% of SwissProt proteins.
Improved orthology prediction and visualization of predicted pathways for model organisms
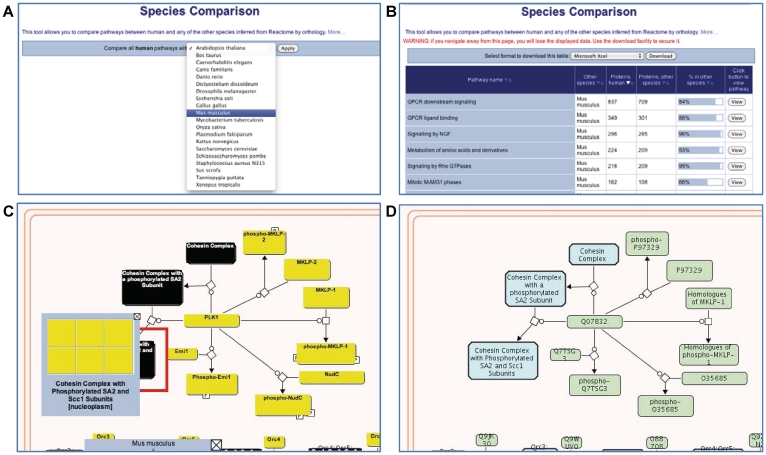
Comparative analysis of biological processes offers important information on their evolution, and supports metabolic engineering, the study of human disease and the identification of potential drug targets. Curated human reactions were used previously to electronically infer reactions by orthology in 20 evolutionary divergent species, with the assistance of the OrthoMCL (25). To align Reactome more closely with the Ensembl set of genome data and genome analysis tools, we have shifted to Ensembl Compara (26) to support orthology-based reaction inferences in 20 species for which high-quality whole-genome sequence data are available, including all 12 of the species in the GO Reference Genome annotation project (27). Viewing diagrams for predicted pathways in another species are available from within the Pathway Browser (Figure 2). A new Species Comparison tool allows users to compare these predicted pathways with those of human to find reactions and pathways common to a selected species and human (Figure 2).
Figure 2.
Species comparison tool and model organism pathway diagrams. (A) A drop-down menu is used to select the model organism species. (B) Results for the comparison of human and mouse pathways. Each row in the table is a pathway; the columns are pathway name, model organism name, number of proteins in the human pathway, number of orthologous proteins in the inferred model organism pathway, a graphical representation of the ratio of these two values and a ‘View’ button that creates a pathway diagram. ‘Sort’ buttons at the top of each column allow the table to be re-ordered according to cell contents in that column. (C) The pathway browser displaying the comparison of the human and mouse Mitotic M-M/G1 pathways. Physical entities in the pathway diagram are color-coded: gray, no match; black, a complex (multicomponent) entity; and yellow, the protein’s ortholog is present in human. (D) The pathway browser displaying the inferred mouse Mitotic M-M/G1 pathway.
Upgraded pathway and expression analysis
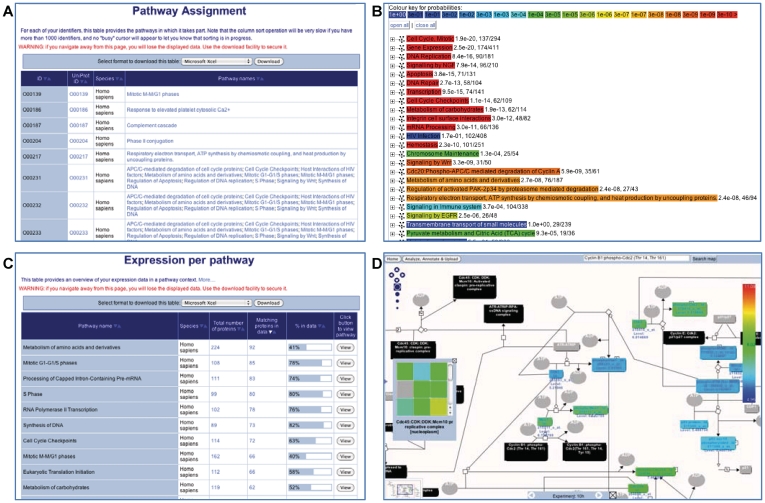
Biologists are generating large amounts of functional data through gene expression, copy-number variation, protein–protein, protein–DNA and protein–RNA interactions, protein and metabolite abundance and large-scale DNA-sequencing experiments. Integrating this experimental information with the published literature and biological databases, including pathway databases, is vital to efficient and effective data analysis. Previously, Reactome provided the Skypainter tool for this level of functional data analysis (1). However, with the retirement of the ‘starry-sky’ reaction map and user requests to provide an expanded suite of bioinformatics tools, we redeveloped our data analysis suite to offer powerful and complementary tools. The Pathway Analysis tool analyzes user-supplied lists of genes, proteins and small molecules and provides ID mapping, pathway assignment and overrepresentation analysis (Figure 3). As with Skypainter, the pathway and expression analysis tools accept gene and protein accession numbers and identifiers that are associated with popular commercial platforms (e.g. Illumina, Agilent and Affymetrix). By default, the simplest of these analyses, ID mapping and pathway assignment, is selected. This analysis takes a set of identifiers and maps them to Reactome pathways. The results are presented in a tabular format (Figure 3). The overrepresentation analysis is based upon the previously reported Skypainter tools. Both pathway analysis results also link to the new Pathway Browser. The expression analysis tool is similar in design to pathway analysis tool, but it will accept numerical values (e.g. expression, abundance, fold change or statistical values) and shows how expression/abundance levels affect reactions and pathways in living organisms (Figure 3). Again, the results are provided in a tabular format. Results from both the pathway and expression analysis results can be downloaded as a spreadsheet or tab- and comma-separated formats. The colored pathway diagrams can be downloaded in publication quality format. The molecular overlay and context sensitive menu features are also enabled in the colored pathway diagrams, providing links from user-supplied experimental data to Reactome pathways and MIs and networks.
Figure 3.
The pathway and expression analysis tools. (A) The results table for the ‘ID mapping and pathway assignment’. The sortable table contains one row for each Reactome pathway and four additional columns: repeats the user-supplied IDs, the corresponding UniProt ID, the species name and names of pathways in which this ID can be found. The names/IDs in the last two columns are clickable links that take the user to a diagram of the named pathway. (B) The results for the ‘overrepresentation analysis’, presented as a list of clickable links of enriched events. The warmer the color, the higher the level of overrepresentation in the given pathway. Clicking on the ‘+’ next to the pathway name gives access to the user-supplied identifiers that are found in the pathway, along with the corresponding UniProt IDs. (C) The results table for the expression analysis. The sortable table contains one row for each Reactome pathway and five additional columns: name of the pathway, Species of presented results, total number of proteins in pathway, number of proteins in the user-supplied data that fall into the pathway, graphical representation of the ratio of these two values and a ‘View’ button that creates a pathway diagram. (D) The pathway browser displaying the colored physical entities that correspond to expression values of the experimental data. The nodes in this diagram have a special color-coding: gray, no match; black, a (multicomponent) complex entity; and other colors represent expression levels. If the numerical data are a time series, the grey bar at the bottom of the colored pathway diagram allows the user to step through time points and visualize changes in expression levels with time of the individual genes involved in the pathway.
Other collaborations and data exports
In collaboration with NCBI, Reactome annotations of pathways are being deposited into the NCBI BioSystems database, a large data repository for cataloguing molecules (nucleic acids, proteins, small molecules, drugs, etc.) that interact in biological systems (28). Reactome is part of the BioPAX Consortium to develop a data-exchange language to describe pathways, reactions and interactions (29). We have partnered with Gene Set Enrichment Analysis (GSEA) group at the Broad Institute to expand the collection of curated gene sets in the Molecular Signatures Database (MSigDB) to include Reactome’s high-quality pathway data (30). Reactome participated in this year’s Google Summer of Code program, collaborating with WikiPathways (31). The integration of pathway and interaction data has been a key element of the Reactome redevelopment. We have provided a new file format for the exchange of binary interaction data, based upon the PSI-MITAB format (32). In response to user requests, we recently changed the representation of protein modifications in Reactome to the PSI-MOD standard (33). We continue to support the use of Reactome data for ontology development with our relationships with the Gene and Protein Ontology groups. Reactome web pages link out to many online bioinformatics databases. This year, additional cross-references to RSCB Protein Data Bank (34), Comparative Toxicogenomics Database (35), DockBlaster (36), BioGPS (37) and dbSNP (38) have been added to the protein pages. Reactome software and data are now distributed under the terms of a Creative Commons Attribution 3.0 Unported License, that grants parties the non-exclusive right to use, distribute and create derivative works based on Reactome, provided that the software and information is correctly attributed to CSHL, OICR and EBI.
FUNDING
National Human Genome Research Institute at the National Institutes of Health (grant number P41 HG003751); European Union 6th Framework Programme ‘ENFIN’ (grant number LSHG-CT-2005-518254). Funding for open access charge: Ontario Institute for Cancer Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to the many researchers who have volunteered to be external authors and reviewers. Development of the Reactome data model and fly and chicken databases is a collaborative project and this work benefited greatly from our interactions with Carl Schmidt, Mark Williams and Michael Ashburner.
REFERENCES
- 1.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ontology Consortium Gene. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degtyarenko K, Hastings J, de Matos P, Ennis M. ChEBI: an open bioinformatics and cheminformatics resource. Curr. Protoc. Bioinform. 2009 doi: 10.1002/0471250953.bi1409s26. Chapter 14: Unit 4.9. [DOI] [PubMed] [Google Scholar]
- 4.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jassal B, Jupe S, Caudy M, Birney E, Stein L, Hermjakob H, D’Eustachio P. The systematic annotation of the three main GPCR families in Reactome. Database. 2010 doi: 10.1093/database/baq018. 2010, doi:10.1093/database/baq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahlem P, Clegg A, Reisinger F, Xenarios I, Hermjakob H, Orengo C, Birney E. ENFIN–A European network for integrative systems biology. C. Roy. Biol. 2009;332:1050–1058. doi: 10.1016/j.crvi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Le Novere N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 13.Killcoyne S, Carter GW, Smith J, Boyle J. Cytoscape: a community-based framework for network modeling. Methods Mol. Biol. 2009;563:219–239. doi: 10.1007/978-1-60761-175-2_12. [DOI] [PubMed] [Google Scholar]
- 14.Funahashi A, Jouraku A, Matsuoka Y, Kitano H. Integration of CellDesigner and SABIO-RK. In Silico Biol. 2007;7:S81–S90. [PubMed] [Google Scholar]
- 15.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2009;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overington J. ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J. Comput. Aided Mol. Des. 2009;23:195–198. doi: 10.1007/s10822-009-9260-9. [DOI] [PubMed] [Google Scholar]
- 20.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reference Genome Group of the Gene Ontology Consortium. The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D’Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 31.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montecchi-Palazzi L, Beavis R, Binz PA, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS. The PSI-MOD community standard for representation of protein modification data. Nat. Biotechnol. 2008;26:864–866. doi: 10.1038/nbt0808-864. [DOI] [PubMed] [Google Scholar]
- 34.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 35.Wiegers TC, Davis AP, Cohen KB, Hirschman L, Mattingly CJ. Text mining and manual curation of chemical-gene-disease networks for the comparative toxicogenomics database (CTD) BMC Bioinformatics. 2009;10:326. doi: 10.1186/1471-2105-10-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y. Automated docking screens: a feasibility study. J. Med. Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]