Abstract
The RCSB Protein Data Bank (RCSB PDB) web site (http://www.pdb.org) has been redesigned to increase usability and to cater to a larger and more diverse user base. This article describes key enhancements and new features that fall into the following categories: (i) query and analysis tools for chemical structure searching, query refinement, tabulation and export of query results; (ii) web site customization and new structure alerts; (iii) pair-wise and representative protein structure alignments; (iv) visualization of large assemblies; (v) integration of structural data with the open access literature and binding affinity data; and (vi) web services and web widgets to facilitate integration of PDB data and tools with other resources. These improvements enable a range of new possibilities to analyze and understand structure data. The next generation of the RCSB PDB web site, as described here, provides a rich resource for research and education.
INTRODUCTION
The RCSB Protein Data Bank (RCSB PDB) (http://www.pdb.org) (1) is a member of the Worldwide Protein Data Bank (http://www.wwpdb.org) (2). The wwPDB partners RCSB PDB (USA), PDBe (Europe, http://pdbe.org) (3), PDBj (Japan, http://www.pdbj.org) and BMRB (USA, http://www.bmrb.wisc.edu) act as data deposition, processing and distribution centers for PDB data. The PDB archive is the single world-wide repository of experimentally determined structures of proteins, nucleic acids, and complex biomolecular assemblies that is curated and annotated following standards set by the wwPDB (4). Each wwPDB partner offers unique views, query, analysis and visualization tools, and web services for the PDB archive on their respective web sites and databases. The RCSB PDB web site has undergone significant changes to improve usability, provide new query and analysis features, integrate additional external resources and enable user customization of the resource. In the 5 years since our last major report (5), the user base has increased from ∼120 000 unique users (based on number of unique IP addresses) per month to ∼180 000 unique users per month. At the same time, the archive has doubled from around 34 000 entries at the end of 2005 to almost 68 000 structures as of September 2010. RCSB PDB web site development has required a scalable infrastructure to accommodate the rapid growth of the archive, increased size and complexity of the data, and an expanding and broadening user base. The RCSB PDB web site caters to a wide variety of ‘customers’ from education (K-12, undergraduate, graduate), to academic and industrial researchers, to programmers and web developers. The redesigned web site supports the disparate requirements of this diverse user base.
Here, we describe major new or expanded features from those reported 5 years ago (5), including new query and analysis tools, options to customize the web site, structural comparison of representative protein chains in the PDB, integration with literature from PubMed Central (http://www.ncbi.nlm.nih.gov/pmc) and binding affinity data from BindingDB (http://www.bindingdb.org). For web developers, we describe new RESTful web services and web widgets which enable the integration of RCSB PDB services and data into other web resources.
QUERY AND ANALYSIS
Chemical components search
About 70% of PDB structures contain ligands such as small molecules, ions, non-aqueous solvents and standard and modified amino acids and nucleotides, collectively referred to as chemical components. The Chemical Component Dictionary (http://www.wwpdb.org/ccd.html) contains the unique set of all components in the PDB (∼11 000 entries). A rich user interface provides the following search options.
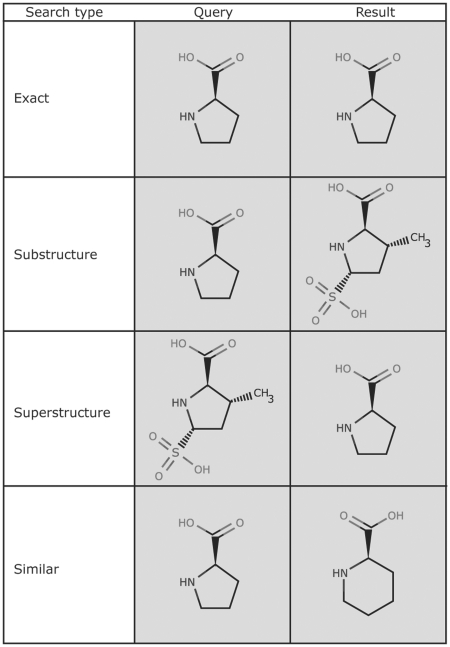
The ‘Structure’ query of the ‘Chemical Components’ Search performs chemical structure searches using SMILES (6) and SMARTS (http://www.daylight.com) linear notations. Search types include exact match, substructure, superstructure and similarity (Figure 1). Similarity searches are based on the Tanimoto coefficient as implemented in ChemAxon JChem Base (http://www.chemaxon.com). Alternatively, chemical structures are drawn with the ChemAxon MarvinSketch Java applet. To facilitate structure drawing, chemical components (ligands) can be loaded by ‘3-letter’ code or SMILES string or imported by name (systematic or common) and further modified. All advanced query features of MarvinSketch are supported, including generic query atoms, atom lists and ‘any’ bonds.
Figure 1.
The Chemical Component Search interface supports four search types. The query structure is shown on the left and an example structure that matches the query is shown on the right. The exact, substructure and superstructure search match the specified stereochemistry, if it is specified in the query. The similarity search ignores stereochemistry.
The ‘Name/Identifier’ query of the ‘Chemical Components’ Search supports searches using the chemical component ID, the InChI string and InChI key (http://www.iupac.org/inchi), and the chemical name. A ‘sounds like’ feature finds chemical component with slightly misspelled chemical names.
The ‘Formula/Weight query’ of the ‘Chemical Components Search’ offers simple and advanced molecular formula searches. Chemical element wildcard ranges and excluded elements can be specified in molecular formulas. For example the expression ‘C5-10 N* O2 P0’ specifies compounds with 5–10 carbon atoms, one or more nitrogen atoms, two oxygen atoms and no phosphorus atoms. A powerful formula expression editor can be launched to compose complex formula queries. This feature is useful for creating ligand sets with a given composition and molecular weight ranges.
Chemical component queries are accessible from the ‘Advanced Search’ menu and can be combined with any other ‘Advanced Search’ options. For example, a substructure search can be combined with a molecular weight range and EC number search to find inhibitors for a specific class of enzymes. Various display options are available for query results, such as tabular reports, and chemical components can be exported in .sdf file format. The next section describes further how query results can be refined.
Query refinement through data drill-down
For some time, the RCSB PDB web site offers two distinct strategies to find information and structures, search and browse. The search from the top bar of the site performs keyword, author, chemical component, and PDB ID searches. For more specific searches, the ‘Advanced Search’ interface offers queries for different categories, including keywords and database identifiers, sequence and structural features and annotations and experimental method details. On the other hand, database browsers present a hierarchical organization of the PDB entries by categories such as ontology terms Gene Ontology (GO) (7) and Medical Subject Headings MeSH (http://www.nlm.nih.gov/mesh), Enzyme commission EC number (http://www.chem.qmul.ac.uk/iubmb/enzyme), source organism (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html) or domain annotations SCOP (8) and CATH (9). With the latter approach, the user can start with a general category and traverse down the hierarchy until a suitable subcategory is found.
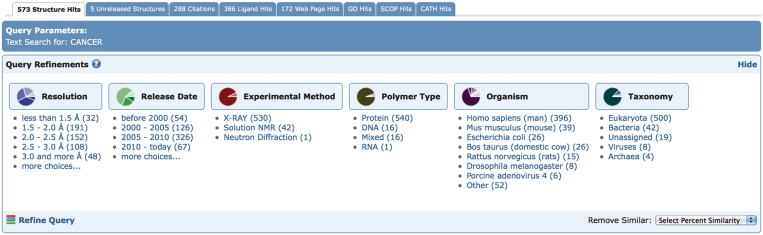
A recently implemented third-search strategy provides faceted navigation (10) by combining searching and browsing functionalities. Often a user is interested in querying the PDB for a particular protein. However, an initial text search by protein name or protein sequence may results in tens or hundreds of hits. To aid the user in analyzing the result set, we display the distribution of the hits by various criteria or facets (Figure 2), which were chosen based on frequently asked questions by users. After analyzing the distribution, the user may pick a category and drill-down further to a subset of the results. In an interactive and iterative process, the user navigates to a subset of interest. The advantage of this approach is that loosely defined queries are refined by using information discovered during the search process. Many e-commerce sites have adopted these hierarchical faceted search interfaces for browsing catalogs of items. Each category can be drilled-down further, exposing more details. For example, a user having completed a keyword search now wants to retrieve the subset of human proteins that match the keyword. By selecting Homo sapiens from the organism category, a new subcategory is displayed. This may include structures that contain only human proteins, or may contain structures that have components from multiple organisms including human. By drilling down on H. sapiens, the structures are further subdivided into pure H. sapiens and mixed cases, for example H. sapiens/Mus musculus, where a structure contains both human and mouse components. The query can now be further refined by selecting subcategories from other categories, e.g. the option polymer type can be used to select proteins and exclude nucleic acid-containing structures. Furthermore, a user can define custom data ranges for numerical values such as resolution or release date. A query description shows the path of the query to guide the user through the process, for example starting with a text search, followed by an organism query, and then a final selection by polymer type. The user can go back up a level at any point and change the search criteria.
Figure 2.
The query-refinement-user interface on the query results page displays the distribution of search hits by various categories. The number of hits for each subcategory is displayed in parenthesis and links to the corresponding subset of hits.
The drill-down feature described here is seamlessly integrated with the advanced query system, and allows combinations of drill-downs with other ‘Advanced Search’ criteria to refine a query. Internally, all queries are represented in an XML format. These queries are stored with each user session, and can be recalled, modified, permanently stored in the MyPDB account of a user, or executed through a web service (both described subsequently).
Tabular reports and data export
The ‘Generate Reports’ system is a quick and easy way for users to view and export query results in a tabular format. Summary reports about structure, sequence, ligand, literature and biological details are provided by default. Specialized experimental detail reports are available for X-ray and NMR structures. A custom table can be created by selecting fields from a list, which includes experimental structural and non-structural data, references to sequence databases [UniProtKB (11), Pfam (12)], domain information (CATH, SCOP), literature (PubMed) and ontology terms (GO, MeSH).
Advanced web technology has been utilized to implement a rich user interface for sorting and filtering the tabular data. Search results can be refined within the tabular report by using advanced filter features. The generated report can be exported in Excel and CSV formats. The Excel spreadsheets are preformatted with customized column width, text wrapping, alignment and hyperlinks on selected columns. The scalability of the tabular reports has been improved so that reports for all entries in the PDB can be generated.
WEB SITE CUSTOMIZATION
MyPDB—new structure alerts
MyPDB provides user accounts and the framework for a personalized web site. After creating a user account, queries can be saved from simple keyword searches to ‘Advanced Searches’. These queries can be automatically run weekly or monthly and users will be notified by email when new structures matching the stored queries are released. MyPDB will be expanded to allow further customization of the site, including storage of personalized structure annotations.
Site layout customization
Since the RCSB PDB’s users may only be interested in very specific topics, we have added options to customize the layout of frequently used pages. Throughout the site are web widgets, which are boxes containing specific information that can be repositioned, hidden or shown on a page. The left-hand menu is comprised of these widgets so users can move the preferred boxes to the top, and hide options that are not as important to the user. The content of the home page can be customized by choosing from a list of main and side panels. For example, a bioinformatics scientist may prefer to have the sequence search box on the home page, whereas a structural biologist may want the deposition widget to appear at the top.
Query results are presented in a concise layout, with information about polymer and ligand details, and full abstracts hidden by default. The user can expand the display as needed. The ‘Structure Summary’ page is the single-most-used page on the web site and provides information about a single-PDB entry. Since users have different interests, the information displayed on this page can be set and arranged in the most meaningful way for a given user. Layout changes are currently stored in a cookie on the web browser. When the user returns to the site from the same computer and browser, the customizations are retained.
STRUCTURE COMPARISON AND VISUALIZATION
Pair-wise structural alignments
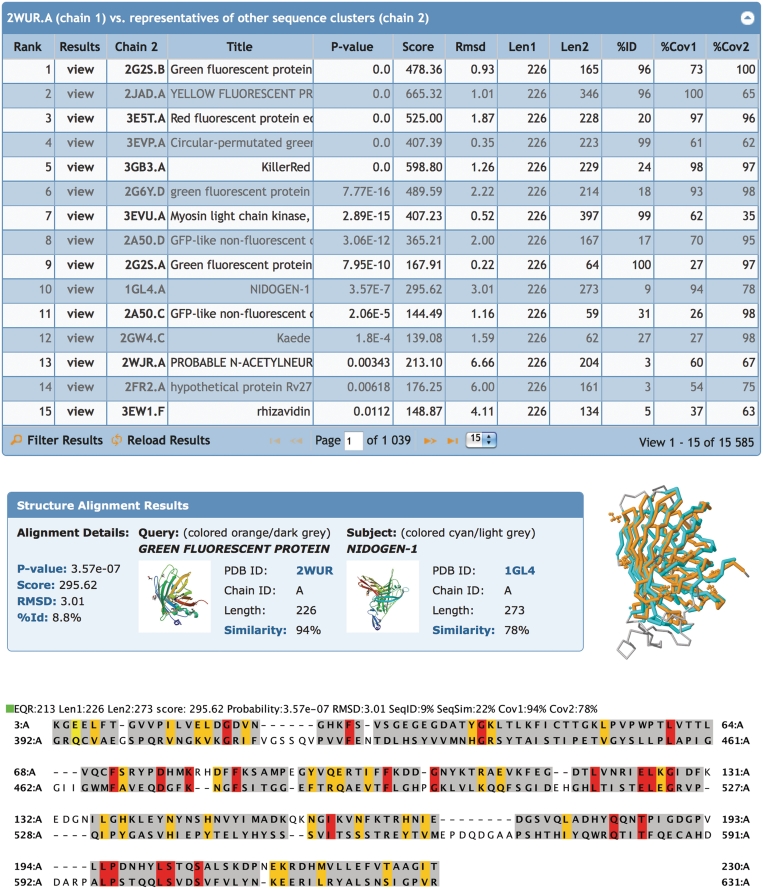
The RCSB PDB’s Comparison Tool calculates pair-wise 3D structure comparisons, as well as sequence alignments. This tool utilizes new Java implementations of the CE (13) and FATCAT (14) structure-alignment algorithms and provides references to external structure alignment servers. A new structure alignment service allows server side calculation of 3D alignments. The tool is complemented by a Java Web Start application that allows custom calculations and provides a novel user interface for the visualization of sequence-3D relationships in the alignment (Figure 3) (15).
Figure 3.
Structural alignments. Top: list of representative protein chains that are structurally similar to green fluorescent protein PDB ID 2WUR, chain A (2WUR.A) calculated by jFATCAT. The list is sorted by P-value: significance of alignment; Score: raw alignment; Cα RMSD: the deviation of the aligned residues; Len1: length of the query chain; Len2: length of subject chain; %ID: percent sequence identity; %Cov1: coverage or % of residues in the query chain that are aligned, %Cov2: coverage or % of residues in the subject chain that are aligned. Middle: structural alignment results for green fluorescent protein 2WUR.A (orange) with nidogen-1 1GL4.A (cyan). Unaligned residues are rendered in grey. Bottom: the sequence alignment shows structurally aligned residues highlighted: identical residues (red), similar residues (yellow), other aligned residues (grey) and unaligned residues (white).
Representative structural alignments
The RCSB PDB uses new tools to provide pre-calculated structural alignments for representative protein chains in the PDB. Representatives are chosen to make the comparisons computationally tractable. The comparison consists of two steps: In the first step, protein sequences are clustered at 40% sequence identity using BLASTClust (http://www.ncbi.nlm.nih.gov/Web/Newsltr/Spring04/blastlab.html). Sequences within a cluster are first ranked by resolution and then by deposition date. The top-ranking sequence of each cluster is taken as a representative for all cluster members under the simplifying assumption that at 40% sequence identity structural features are conserved. In the second step, all pairs of representative protein chains are structurally aligned with the jFATCAT rigid method and stored in a database. The alignments are updated weekly with new incoming protein structures.
Novel domain architectures as well as unexpected structural similarities can be found by analyzing these structural alignments. As an example, the structural alignment results for green fluorescent protein [GFP; PDB ID: 2WUR (16)] are displayed in Figure 3. One of the top-ranking results is nidogen-1. Of GFP (Aequorea victoria) residues, 94% align with the nidogen-1 G2 fragment (M. musculus) with a RMSD of 3 Å based on the Cα positions, but with surprisingly low-sequence identity of ∼9%. Indeed, the structural similarity has been recognized by Hopf (17). Nidogen-1, also known as entactin, is a component of the basement membrane (18) and does not contain a chromophore. PSI-BLAST searches do not recognize the relationship between the two proteins, however, the strong structural conservation of the 11-stranded β-barrel and the internal helices suggest that the nidogen-1 G2 fragment and GFP share a common ancestor (17). This example demonstrates the utility of having pre-calculated structural alignments available.
Visualization of biological assemblies
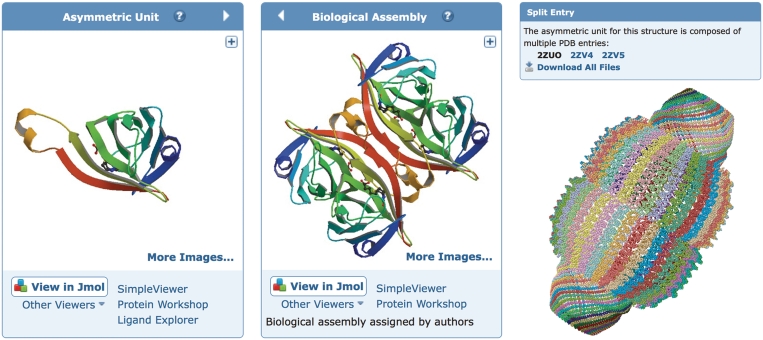
The deposited coordinate set may not always represent the biologically relevant assembly(s). For example, for structures determined by X-ray crystallography, the asymmetric unit is the smallest portion of a crystal structure to which symmetry operations can be applied in order to generate the complete unit cell. On the ‘Structure Summary’ page, we display both the asymmetric unit and the biological assembly(s) (Figure 4), with the latter being the default view. The biological assembly is either specified by the structure author or assigned by PISA (19) or PQS (20) software and manually checked by PDB annotators. The biological assembly is generated from the asymmetric unit by applying the symmetry transformation specified in the PDB entry.
Figure 4.
Structure visualization. Left: the asymmetric unit of streptavidin in PDB entry 1STP. The 3D viewers listed below the image will display the asymmetric unit. The arrow toggles between the asymmetric unit and the biological assembly. Middle: the biological assembly of streptavidin based on oligomeric state assigned by the author. The 3D viewers listed below the image will display the biological assembly. Right: a very large biological assembly, the rat liver vault protein, visualized in the Jmol viewer. The structure is the composite of the three PDB entries 2ZUO, 2ZV4 and 2ZV5 with symmetry transformation (−x, y, −z) applied to generate the complete biological assembly. To accommodate the large size, only the Cα positions are used to render the structure.
Visualization of large assemblies and split entries
Advances in structure determination methods have led to an increase in the size and complexity of biological macromolecules in the PDB. Display of very large assemblies comprising >1 million atoms, or thousands of protein chains, poses a challenge to currently available 3D-visualization software, as well as pushing the memory limits of standard personal computers. Despite the increasing speed of the internet, even the download of these large structures files (>100 Mb) becomes an issue for interactive visualization. We have developed methods to display any large assembly in the PDB on a modern laptop or desktop computer.
Several enhancements to the Jmol (http://www.jmol.org) viewer page have been made to display very large structures. Previously, some structures with very large coordinate files could not be displayed because they would have required more memory than was available to the Jmol applet. We are now able to display structures that contain a large number of chains [e.g. 1GAV (21)], structures that contain very large molecules [e.g. 1JJ2 (22)], structures that contain many models of relatively small macromolecules [e.g. 2HYN (23)] and structures that contain multiple models of large molecules [e.g. 1HTQ (24)]. These structures are loaded into Jmol using a version of the PDB coordinate file that includes only backbone atoms for all polymers (Cα atoms for proteins and P atoms for nucleic acid chains). All modified residues, ligands and water are retained and displayed as well.
A number of structures in the PDB are so large that the historical limitations of the PDB file format (five columns for atom numbering, one column for chain ID) require the structures to be split across multiple PDB coordinate files. These structures include extremely large ribosome complexes [e.g. 1GIX, 1GIY (25)] and other structures that contain a very large number of atoms or chains, such as the vault protein (Figure 4), which is composed of the PDB entries 2ZUO, 2ZV4 and 2ZV5 (26). The images and Jmol view available from the ‘Structure Summary’ page now display the complete structure. The individual entries that comprise the structure are now identified by the new ‘Split Entry’ box on any one of the ‘Structure Summary’ pages for any one of the PDB IDs. This box lists the PDB IDs of all entries that make up the composite structure, and links to the ‘Structure Download’ Tool to easily access the related files in any format. Composite views for both asymmetric unit and biological assemblies are displayed for split entries.
INTEGRATION WITH OTHER RESOURCES
Integration with open access literature
The boundaries between scientific databases and journals are blurring (27). Data are increasingly accessible as supplemental information to the paper, whereas databases are adding curated information taken directly from the literature. To address this trend, we have added a new ‘Literature View’ which, for each structure, reports all open access articles citing or mentioning that particular PDB ID in the full text of the article, as well as a list of related PDB entries that have been mentioned together in the same article(s) (28). For these open access articles taken from PubMed Central (http://www.ncbi.nlm.nih.gov/pmc), a BioLit version (http://biolit.ucsd.edu; 29) of those articles is available which includes semantic markup and references to ontological terms used in those articles. The context in which the PDB ID appears in the full-text article is also given. The overall impact is to bring elements of the literature directly to the RCSB PDB.
Integration with binding affinity data
Structural and energetic data are essential for the understanding of molecular interactions. BindingDB (30) collects binding data for proteins that are either validated or putative drug-targets, and for which the PDB holds representative structural data. BindingDB currently contains ∼500 000 interaction data for ∼3000 protein targets (http://www.bindingdb.org). Integrated structure and affinity data are now available and are particularly valuable to researchers engaged in drug-design projects and to scientists calibrating, benchmarking and validating computational methods for predicting binding affinities.
Binding affinity data including the binding constants IC50, EC50, Ki and thermodynamic data Kd, ΔG°, ΔH°, -TΔS° are exchanged between BindingDB and the RCSB PDB. These data are listed on the ‘Structure Summary’ pages of the corresponding protein–ligand complexes, with links back to BindingDB pages that contain detailed information about the experiment. An ‘Advanced Search’ is available to find structures with associated binding affinities. On the ‘Ligand Summary’ page links to related entries in BindingDB are provided. Conversely, BindingDB maintains links to the RCSB PDB web site for individual protein–ligand complexes, ligands and uses RCSB PDB web services to identify sequences and chemical structures in the PDB that are similar to those in BindingDB, thus providing a structural context to binding-affinity data. The bi-directional links between BindingDB and RCSB PDB enable the correlation of binding-affinity data with appropriate structural data and vice versa.
Web services
RCSB PDB web services provide programmatic access to query tools and PDB data via the Hypertext Transfer Protocol (HTTP). We support both Simple Object Access Protocol (SOAP) and Representational State Transfer (REST) web services. SOAP services have been supported for several years; more recently light-weight RESTful services were added. Future work on web services will only use the RESTful protocol, due to their simplicity. SOAP services will not be developed any further due to their complexity. Here we describe two types of RESTful services (Table 1): (i) ‘Fetch services’ return experimental data based on PDB IDs, entity IDs (PDB IDs + Chain IDs), or Chemical Component (Ligand) IDs passed to the server; (ii) ‘Search services’ perform a query on the PDB database and return results in an XML format.
Table 1.
RESTful web services supported by the RCSB PDB
| Fetch services | PDB entry: description, unique chains (entities), release status. |
| Chemical Components (ligands): description, occurrence in PDB entries. | |
| Sequence and structure clusters and representative structures. | |
| Sequence and domain annotations: GO, UniProt, SCOP, CATH, PFAM, InterPro, DP, PDP, secondary structure. | |
| Search services | Search chemical components by SMILES: exact match, substructure, superstructure, similarity and SMARTS match. |
| Advanced search: any search supported by the RCSB PDB site. |
Detailed descriptions and examples are available at: http://www.pdb.org/pdb/software/rest.do.
RESTful services are easy to use; for example the following URL:
http://www.pdb.org/pdb/rest/describeMol?structureId=4hhb
specifies a fetch service that returns a description in XML format for the polymer entities in PDB entry 4HHB:
<structureId id="4HHB">
<polymerDescriptions>
<polymer entityNr="1" length="141" type="polypeptide(L)">
<chain id="A"/>
<chain id="C"/>
<polymerDescription description="HEMOGLOBIN (DEOXY) (ALPHA CHAIN)"/>
</polymer>
<polymer entityNr="2" length="146" type="polypeptide(L)">
<chain id="B"/>
<chain id="D"/>
<polymerDescription description="HEMOGLOBIN (DEOXY) (BETA CHAIN)"/>
</polymer>
</polymerDescriptions>
</structureId>
This information can be easily parsed with an XML parser and used by an application or web site.
Web widgets
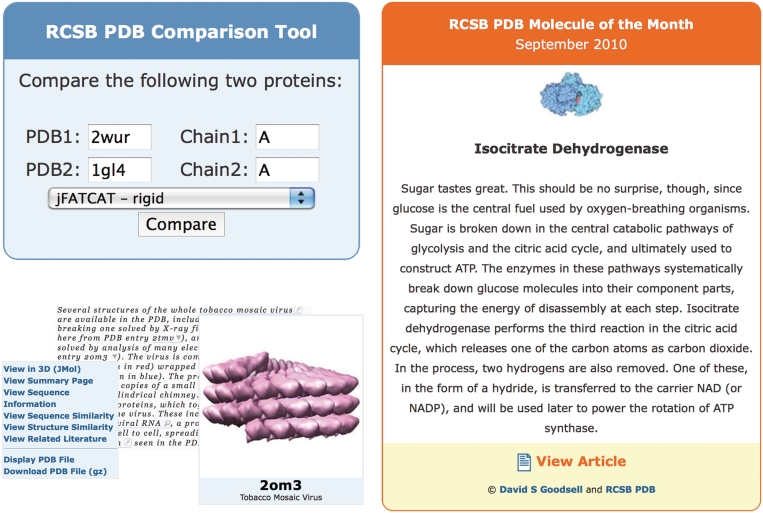
Third-party web sites often display PDB-related data. Traditionally, another resource would download or link to information from the RCSB PDB and then display that information on their web site. This approach is not very scalable and requires a constant update by those sites as new entries are added to the PDB. Recognizing this problem, we have created web widgets, small bits of code which provide access to RCSB PDB functionality (31). The widgets are self-contained JavaScript files that we maintain, thereby addressing both scalability and data currency issues; data will be automatically synchronized with the most recent updates. Widgets are customizable; for example, the size and color can be set to match a third party’s web site’s color scheme. An example of extensive use of RCSB PDB widgets can be found at The Open Protein Structure Annotation Network (TOPSAN; http://www.topsan.org), which uses the Molecule of the Month widget on their home page, the Tag Library Widget on each of their web pages and the Comparison Tool widget to find structures in the PDB related by sequence or structural similarity. The Transporter Classification Database (TCDB; http://www.tcdb.org) (32) uses the Image Widget to display images of transport proteins.
Currently, the RCSB PDB offers the following web widgets.
Image library
A widget that embeds a structure image based on a PDB ID. The image size and type of assembly (asymmetric unit or biological assembly) can be customized.
Tag library
A rich mark-up widget that tags PDB IDs and keywords on a web site and automatically provides enhanced functionality that links back to the RCSB PDB web site. Four types of tags are supported by this widget: author tags are used to mark-up author names. For example, structural biologists can use this functionality to provide always up to date links to their published PDB structures on their own web pages. Simple PDB ID tags mark up a section of text or a PDB ID and provide tool tips that display a PDB structure image and link to the ‘Structure Summary’ page. The Menu tag creates a menu to display or download information about a single PDB entry. The Keyword tag marks up a word or phrase of text and links to a query results page.
Comparison tool
A widget that performs the pair-wise sequence and structure alignments between two protein chains described above.
Molecule of the Month (MoM)
A widget that embeds a MoM image and links to the full article. A short paragraph can be optionally displayed. The MoMs are educational articles about important molecules in the PDB. The MoM widget is an ideal way for educational web sites to display the most recent MoM articles. Figure 5 shows examples of these widgets. A detailed description how to use these widgets is available at http://www.pdb.org/pdb/static.do?p=widgets/widgetShowcase.jsp.
Figure 5.
Several widgets are available for embedding PDB resources into third-party web sites. Top left: the comparison tool that may be used to align sequences or structures, shown here in use for a structural alignment of green fluorescent protein and nidogen-1. Bottom left: a PDB ID in a sample document (excerpt from http://www.pdb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb109_1.html) is marked up with the menu tag that displays a menu to view and download information from the RCSB PDB site. Also shown is an example of the image tag for PDB ID 2OM3. Right: the Molecule of the Month widget that presents the first paragraph of the current Molecule of the Month, and links to the full article.
SUMMARY
The RCSB PDB web site continues to take advantage of new scientific understanding and new technological developments. The powerful new Chemical Structure Search interface supports simple molecular weight, formula, substructure or similarity searches and complex SMARTS queries. Faceted navigation significantly improves browsing query results with hierarchical navigation and the ability to refine a query iteratively based on new information gained during the search. Both the Chemical Structure Search and the new faceted search interface are tightly integrated with the ‘Advanced Search’ system to provide multiple paths for query refinement. The results of the queries can be tabulated, sorted, filtered and exported to enable large-scale data analysis.
New sequence and structure analysis tools have been implemented. New Java versions of the FATCAT and CE algorithms for structural superposition have been made available for pair-wise alignments. In addition, all representative protein chains in the PDB have been structurally aligned. These pre-calculated alignments are updated weekly. Novel structures or new folds can be readily identified, as well as unexpected similarities between proteins of low-sequence identity. This information may be used to find evolutionary relationships or deduce previously unknown functions of proteins.
To deal with the visualization of large and complex assemblies, special methods have been created to enable their visualization on standard hardware. File format changes in the future should accommodate the increasing size of molecular assemblies.
PDB entries are now tightly integrated with the open-access literature that uses or cites the entries. The ‘Literature View’ of the RCSB PDB provides new ways to search and analyze the data. Data exchange and bi-directional links with BindingDB enable correlation of structure with binding affinity data. For web developers, we provide web widgets to integrate RCSB PDB tools and data into their web sites, and a RESTful service application user interface (API) enables programmatic access to RCSB PDB queries and data retrieval.
Not all new features and enhancement could be described here. The ‘What’s New’ page (http://www.pdb.org/pdb/static.do?p=general_information/whats_new.jsp) lists detailed summaries of the latest and past improvements. The addition of these new features and improvements represents a new generation of the RCSB PDB web site. It allows more complex analysis to be performed and provides systematic comparisons across all of the PDB with the goal to further scientific discovery and education.
FUNDING
National Science Foundation (NSF DBI 0829586); National Institute of General Medical Sciences (NIGMS); Office of Science, Department of Energy (DOE); National Library of Medicine (NLM); National Cancer Institute (NCI); National Institute of Neurological Disorders and Stroke (NINDS); National Institute of Diabetes & Digestive & Kidney Diseases (NIDDK). Computational resources for structural alignments are provided in part by the Open Science Grid (http://www.opensciencegrid.org) funded by the National Science Foundation; and the Office of Science, Department of Energy (DOE) (NSF 0753335). The RCSB PDB is managed by two members of the RCSB: Rutgers and UCSD. Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to ChemAxon (http://www.chemaxon.com) for providing Marvin Sketch, JChem Base and Standardizer for the chemical-structure search. Michael Gilson and Tiquing Liu at UCSD worked with us on the integration with BindingDB. We thank Sean Van Tyne and the San Diego User Experience Special Interest Group (http://www.uxsig.org) for an in-depth usability review of the RCSB web site that led to many improvements. In addition, we appreciate all users who provided feedback. Finally, we thank other RCSB PDB staff past and present for suggestions, critical review and testing of new features.
REFERENCES
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 3.Velankar S, Best C, Beuth B, Boutselakis CH, Cobley N, Sousa Da Silva AW, Dimitropoulos D, Golovin A, Hirshberg M, John M, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2010;38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the Worldwide Protein Data Bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 5.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 7.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreeva A, Howorth D, Chandonia J-M, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuff A, Redfern OC, Greene L, Sillitoe I, Lewis T, Dibley M, Reid A, Pearl F, Dallman T, Todd A, et al. The CATH hierarchy revisited-structural divergence in domain superfamilies and the continuity of fold space. Structure. 2009;17:1051–1062. doi: 10.1016/j.str.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hearst MA. Search User Interfaces. New York: Cambridge University Press; 2009. [Google Scholar]
- 11.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 14.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19 (Suppl. 2):II246–II255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 15.Prlić A, Bliven S, Rose PW, Bluhm WF, Bizon C, Godzik A, Bourne PE. Precalculated protein structure alignments at the RCSB PDB website. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq572. doi:10.1093/bioinformatics/btq572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinobu A, Palm GJ, Schierbeek AJ, Agmon N. Visualizing proton antenna in a high-resolution green fluorescent protein structure. J. Am. Chem. Soc. 2010;132:11093–11102. doi: 10.1021/ja1010652. [DOI] [PubMed] [Google Scholar]
- 17.Hopf M, Göhring W, Ries A, Timpl R, Hohenester E. Crystal structure and mutational analysis of a perlecan-binding fragment of nidogen-1. Nat. Struct. Biol. 2001;8:634–640. doi: 10.1038/89683. [DOI] [PubMed] [Google Scholar]
- 18.Chung AE, Durkin ME. Entactin: structure and function. Am. J. Respir. Cell Mol. Biol. 1990;3:275–282. doi: 10.1165/ajrcmb/3.4.275. [DOI] [PubMed] [Google Scholar]
- 19.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Bio. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 21.Tars K, Bundule M, Fridborg K, Liljas L. The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses. J. Mol. Biol. 1997;271:759–773. doi: 10.1006/jmbi.1997.1214. [DOI] [PubMed] [Google Scholar]
- 22.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potluri S, Yan AK, Chou JJ, Donald BR, Bailey-Kellogg C. Structure determination of symmetric homo-oligomers by a complete search of symmetry configuration space, using NMR restraints and van der Waals packing. Proteins. 2006;65:203–219. doi: 10.1002/prot.21091. [DOI] [PubMed] [Google Scholar]
- 24.Gill HS, Pfluegl GM, Eisenberg D. Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation. Biochemistry. 2002;41:9863–9872. doi: 10.1021/bi020254s. [DOI] [PubMed] [Google Scholar]
- 25.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka H, Kato K, Yamashita E, Sumizawa T, Zhou Y, Yao M, Iwasaki K, Yoshimura M, Tsukihara T. The structure of rat liver vault at 3.5 angstrom resolution. Science. 2009;323:384–388. doi: 10.1126/science.1164975. [DOI] [PubMed] [Google Scholar]
- 27.Bourne PE. Will a biological database be different from a biological journal? PLoS Comput. Biol. 2005;1:179–181. doi: 10.1371/journal.pcbi.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prlić A, Martinez MA, Dimitropoulos D, Beran B, Yukich BT, Rose PW, Bourne PE, Fink JL. Integration of open access literature into the RCSB Protein Data Bank using BioLit. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-220. 220, doi:10.1186/1471-2105-11-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fink JL, Kushch S, Williams PR, Bourne PE. BioLit: integrating biological literature with databases. Nucleic Acids Res. 2008;36:W385–W389. doi: 10.1093/nar/gkn317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourne PE, Beran B, Bi C, Bluhm W, Dunbrack R, Prlić A, Quinn G, Rose P, Shah R, Tao W, et al. Will widgets and semantic tagging change computational biology? PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000673. e1000673, doi:10.1371/journal.pcbi.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]