Abstract
Allostery is the most direct, rapid and efficient way of regulating protein function, ranging from the control of metabolic mechanisms to signal-transduction pathways. However, an enormous amount of unsystematic allostery information has deterred scientists who could benefit from this field. Here, we present the AlloSteric Database (ASD), the first online database that provides a central resource for the display, search and analysis of structure, function and related annotation for allosteric molecules. Currently, ASD contains 336 allosteric proteins from 101 species and 8095 modulators in three categories (activators, inhibitors and regulators). Proteins are annotated with a detailed description of allostery, biological process and related diseases, and modulators with binding affinity, physicochemical properties and therapeutic area. Integrating the information of allosteric proteins in ASD should allow for the identification of specific allosteric sites of a given subtype among proteins of the same family that can potentially serve as ideal targets for experimental validation. In addition, modulators curated in ASD can be used to investigate potent allosteric targets for the query compound, and also help chemists to implement structure modifications for novel allosteric drug design. Therefore, ASD could be a platform and a starting point for biologists and medicinal chemists for furthering allosteric research. ASD is freely available at http://mdl.shsmu.edu.cn/ASD/.
INTRODUCTION
Allostery, namely allosteric regulation, describes the regulation of protein function, structure and/or flexibility induced by the binding of a ligand at a site topographically distinct from the orthosteric site (1). Such site is then defined as an allosteric site. With growing collection of genome sequences and gene expression profiles, increasing attention has been focused on protein function and regulation in the post-genomic era (2). Allostery is the most direct, rapid and efficient way to regulate protein function, ranging from the control of metabolic mechanisms to signal-transduction pathways (3). Allosteric behaviors are mostly found by the specific binding of metal ions or molecules, which can alter cellular responses in order to maintain homeostasis (1). Dysregulations of allosteric systems are significantly associated with human diseases, such as Alzheimer’s disease, inflammation and diabetes (4–6).
The first cooperativity regulation was observed from the sigmoidal-binding curve of hemoglobin to O2 in 1903 and published in 1910 (7). The remarkable phenomenon has aroused widespread concerns and led to the appearance of the concept of ‘allosteric’ by Jacob and Monod (8,9). Allosteric enzymes were first summarized in the book of Kurganov in 1978 (10), which collected a large amount of experimental information and became a major allosteric reference source. The allosteric family has now expanded from multimeric proteins to monomeric proteins as well as from native proteins to engineered proteins (11–13). Intrinsically, the allosteric effect in a protein transmits conformation change from the allosteric site to the orthosteric site via atom fluctuations, amino acid residue networking or domain motion according to the distance between the sites, eventually leading to the switch of functions between two or more conformational states. A persistent conformation fixed by external factors is able to function sustainably in the state (1).
A common factor for allosteric regulation derives from the binding of metal ions and small molecules to the allosteric sites as allosteric modulators, including activator/agonist, inhibitor/antagonist and other effector types (see below) (1). Chemical allosteric modulators boasts several advantages over orthosteric ligands as potential therapeutic agents due to their quiescence in the absence of endogenous-orthosteric activity, greater selectivity as a result of higher sequence divergence in allosteric site and limited positive or negative cooperation imposing a ‘ceiling’ on the magnitude of their allosteric effect (14). In recent years, remarkable progress has been made in the discovery, optimization and clinical development of allosteric drugs of kinases, GPCRs and ion channels by the pharmaceutical industry; for example, the development of Gleevec (allosteric inhibitor of Abl) (15), Cinacalcet (allosteric activator of calcium sensing receptor) (14) and Maraviroc (allosteric inhibitor of chemokine receptor 5) (14) promises exciting therapeutic prospects with fine regulation and fewer off target side effects.
Despite its significance and usefulness, an enormous amount of unsystematic allostery information has deterred scientists who could benefit from this field. Specialized databases and analysis systems dedicated to allostery are becoming crucial for capturing and describing a rapidly increasing population of allosteric molecules and for better understanding the mechanisms of allosteric proteins and designing allosteric modulators for drug discovery. In this work, we have developed the AlloSteric Database (ASD), a comprehensive database of allosteric proteins and their modulators. This is the first online database, to our knowledge, that focuses on exhaustive allostery information describing the specific structure, function and mechanism of 336 allosteric proteins and 8095 allosteric modulators, together with their statistical evaluation, references to the scientific literature and cross-links to other databases, such as PubMed, UniProt (16), GenBank (17), Enzyme Nomenclature (18), KEGG (19), PDB (20), SCOP (21) and CATH (22). Furthermore, BLAST search engine for proteins and chemical structure search engine for small modulators are available as web-based tool for allosteric recognition. Taken together, ASD is an integrated resource that could provide useful information and tool for the investigation of allosteric mechanism as well as novel drug design and protein engineering.
MATERIALS AND METHODS
Information on allostery was collected from scientific literature and various web resources: e.g. IUPARM (23), Drugbank (24) and PDB (20). Some information was gathered from United States Patent and European Patent files. First, 16 425 abstracts of PubMed were automatically filtered for relevant articles using ‘alloster*’ as keyword. The names of allosteric proteins were then extracted from the abstracts to clusters by a protein name dictionary constructed from UniProt (16), retrieving ∼500 distinct biological proteins. A team of scientists manually processed the papers with respect to the clustered names. With at least three cases of experimental evidence in crystal structure complex or biochemistry (inactive mutation of allosteric residue, cooperativity of kinetic effect from two ligands and uncompetitive-binding assay with chromatography, etc.), 336 proteins supporting their functional change elicited by modulator binding at a site that was topographically distinct from the orthosteric functional site, were verified as allosteric proteins for deposition into the ASD. All proteins in the ASD were annotated with gene information, biological function, natural mutations and related diseases extracted from GenBank (17), Uniprot (16), Enzyme Nomenclature (18) and original literature. An up-to-date synchronization on available structures of allosteric proteins from PDB is present and their structural classification SCOP (21) and CATH (22) based on the PDB ID are also labeled. Large-scale allosteric conformational changes, such as open–close and twist, inferred from multiple conformational structures of allosteric proteins, were manually annotated to the allosteric mode field. Theoretical models of allosteric proteins without structures were generated with I-TASSER (25) or built manually using Modeller (26) when C-score of the best I-TASSER model is below −1.5 or high-homologous oligomeric templates are available in PDB. All structures are downloadable as PDB files. Notably, extensive descriptions of allosteric sites were summarized from literature and the sites were always highlighted in the diagram of protein topologies (27,28) if they have been explicitly validated by biochemistry or structure biology.
Second, after all allosteric proteins with relevant annotation information were collected, we further searched for allosteric modulators for the 336 allosteric proteins. All the abstracts from PubMed, United States Patent and European Patent files containing ‘allosteric modulator/effector/activator/inhibitor/agonist/antagonist’ in combination with the name of the collected allosteric proteins were curated and then manually identified as the final set, resulting in the collection of 8095 chemical allosteric modulators with respective references. Meanwhile, publicly available binding affinities of the allosteric modulators to their allosteric sites in the proteins were also obtained from the references. Among the allosteric modulators in the ASD, those that increase a particular protein function, for example, orthosteric ligand affinity or catalytic rate, are classified as ‘allosteric activator’ or ‘A’. Those that decrease a particular protein function are classified as ‘allosteric inhibitor’ or ‘I’. The remaining modulators, which control cooperativity of multimeric proteins, regulate the enzyme indirectly under allosteric control via the bound protein with an allosteric site, induce a new binding site to a protein, etc., are classified to the ‘allosteric regulator’ or ‘R’ category. Since allosteric modulators were initially identified from endogenous ligands and then widely accepted for the development of novel types of drugs, the tags ‘Endogenous’ and ‘Druggable’ in the ASD differentiate allosteric modulators produced in vivo and designed for drug use, respectively. In addition, important physicochemical properties used in drug discovery, such as logP, PSA, the number of rotatable bonds, etc., were calculated on the allosteric modulators by Filter Program from Openeye (http://www.eyesopen.com). Each modulator in the ASD is downloadable as 2D mol and 3D mol2 files.
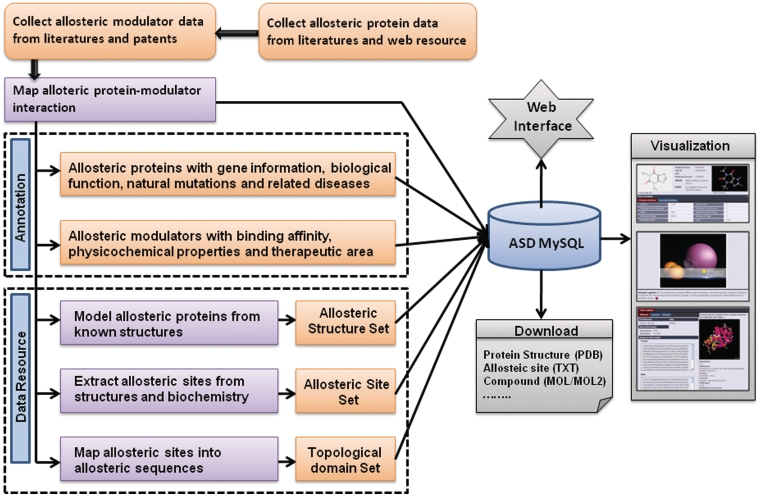
ASD is designed as a relational database on a MySQL server (Figure 1). For visual inspection of 2D chemical structures, MarvinView (http://www.chemaxon.com) is installed. For displaying 3D structures, Jmol (http://www.jmol.org/)—an open source Java viewer for proteins and chemical structures in 3D—is used. MarvinSketch is applied as a built-in molecule editor, which allows users to query the database using self-edited molecules. The web site is built with Ajax framework ExtJS with EJB3, Web access is enabled via JBOSS Webserver. Internet Explorer version 7 or above, Mozilla Firefox version 3.6 or above, Apple Safari and Google Chrome were thoroughly tested and thus recommended for ASD.
Figure 1.
The basic framework of ASD. All results generated by ASD are deposited in MySQL and displayed in the visual browser and web pages. The web-interface programs and browser can be accessed by a wide range of research biologists and chemists to analyze and visualize data over the internet.
RESULT
The ASD database
ASD is a comprehensive database on allosteric information. It contains two types of data: (i) allosteric proteins; (ii) small allosteric modulators identified from in vitro binding to the allosteric proteins at the allosteric site. In total, 336 allosteric proteins and 8095 chemical allosteric modulators are curated and fully annotated by the database developers and experts in the field.
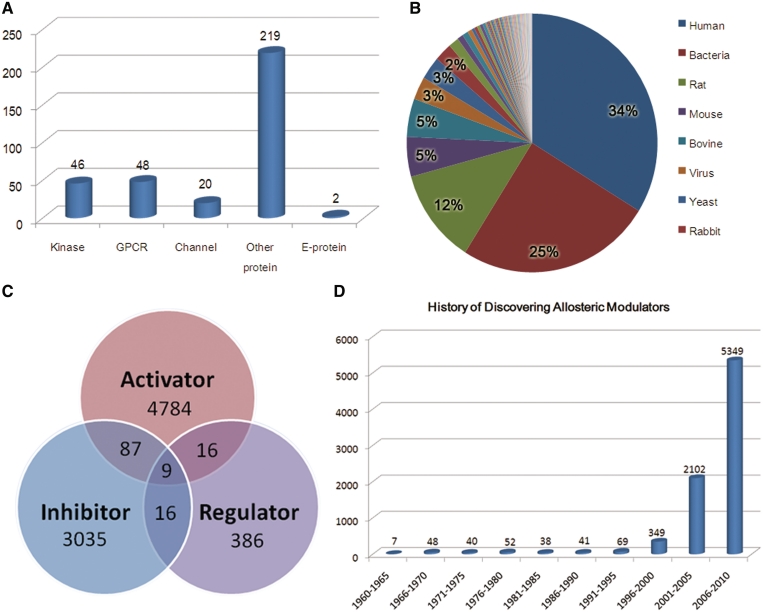
Allosteric proteins in ASD cover 101 different species, of which 34% of the allosteric proteins are from human and 25% from bacteria (Figure 2B). Crystal structures of 3580 redundant proteins are provided from PDB and 156 of them explicitly show allosteric sites. Based on the known structures, theoretical 3D models of the remaining 220 missing allosteric structures from 50 species are constructed and downloadable from ASD. We have analyzed the occurrence of allosteric proteins in the structural hierarchies represented in SCOP and CATH. Class with mainly ‘alpha/beta’ in SCOP and ‘mixed’ in CATH are largely overrepresented in the allosteric proteins, which indicates that secondary structures of a certain scale are important in allostery. More and more allosteric proteins have been revealed to be disease causing when abnormal and some of them used as drug targets. Therefore, 248 diseases from irregular allostery are also included in ASD.
Figure 2.
Statistics on the allosteric proteins and modulators. (A) Class distribution of allosteric proteins. (B) Species distribution of allosteric proteins. (C) Category distribution of allosteric modulators. (D) A history of the discovery of allosteric modulators.
Currently, ASD contains 8095 allosteric modulators. Of them, 4784 activators, 3035 inhibitors and 386 regulators were verified in literature to be associated with corresponding allosteric proteins, and only 101 modulators (1.2%) play multiple roles in different allosteric systems as shown in Figure 2C. Thus, 8680 allosteric interactions between proteins and modulators are deposited into ASD. An overwhelming portion of the modulators is organic small molecules (8062, 99.6%), followed by ions (27, 0.33%) and peptides (6, 0.07%). Since almost all allosteric hits/leads/drugs were initially derived from endogenous molecules and then screened and modified in drug discovery, ASD now holds 244 endogenous allosteric seeds and 7481 compounds in the pipeline of drug discovery.
Web interface
ASD provides a variety of interfaces and graphical visualizations to facilitate viewing and analysis of the allosteric molecules (proteins and modulators) from structures, functions and related properties. As shown in Supplementary Figure S1 in the supporting information, ASD presents the three starting points of browsing ASD and three search options. To visually understand the data in ASD, browsing and searching tools are fully crosslinked. One can quickly jump from search results to their full pages of browsing so that the users can analyze data more efficiently. For example, users can start by searching the name of an allosteric molecule and visualize a complete description in the browsing page and then download the specific molecule for further review.
ASD supports flexible query for various allosteric molecules and related structural and function annotation by providing three ‘Search’ tools—‘Blast search’, ‘Modulator search’ and ‘Text search’. ‘Blast search’ is powered by PSIBLAST (29) and is particularly useful as it allows users to quickly identify allostery by comparing the query proteins to known allosteric proteins in ASD. The search is triggered by pasting a FASTA format sequence and pressing the ‘search’ button, resulting in a list of similar allosteric proteins reported in terms of E-values. A significant hit reveals the possibility that the query protein may act with allostery in a way similar to the allosteric template. In addition, the specific allosteric site in the concerned allosteric protein could be validated by alignment to other family proteins. ‘Modulator search’ can be used to design novel allosteric compounds of known allosteric proteins in ASD. User may sketch (through Marvin’s freely available chemical sketching applet) or paste a SMILES string (30) of a possible allosteric compound into the Modulator search window. Submitting the query launches a structure similarity search tool that looks for common features from the query compound that match known allosteric modulators in ASD. High-score hits are ranked in a tabular format with hyperlinks to the corresponding full description and in turn to links to the allosteric protein target. The ‘Modulator search’ tool allows users to quickly determine whether their compound of interest acts on the desired allosteric protein target and reveal whether the compound of interest may unexpectedly interact with unintended allosteric protein targets. In addition to these structure similarity searches, the Modulator search utility also supports compound searches on the basis of physicochemical properties and chemical formulas. ‘Text search’ provides users a global tool to search throughout ASD by typing a single term, such as a name, a PDB identifier or a species that is related to an allosteric molecule of interest and the server will return a list of links to relevant entries. Each entry contains a brief introduction of the allosteric molecule with a hyperlink to its full page.
The ‘Browse’ tools in the database facilitate easy retrieval of information from ASD through three categories: ‘Modulator browse’, ‘Target browse’ and ‘Index browse’. ‘Modulator browse’ is used to visualize all allosteric modulators with 2D structures and synoptic description in the tab of ‘All’ or three respective categories (‘Activator’, ‘Inhibitor’ and ‘Regulator’) at the first level, in which each entry links to its second level for exhaustive description of interest. The detailed annotation contains name of the molecule, molecular weight, interactive applets for viewing 2D and 3D molecular structures, >30 drug-like physicochemical properties and experimentally validated allosteric targets with hyperlinks to references. This is designed for pharmacists and medicinal chemists who work closely with the quantitative structure–property relationship of allosteric modulator. ‘Target browse’ allows user to preview the list of names of allosteric proteins under class tabs of ‘Kinase’, ‘GPCR’, ‘Channel’, ‘E-protein’ and ‘Other’ at the first level (Figure 2A) and a quick navigation of allosteric protein of interest in all validated species integrated at a panel of the second level is shown by clicking on the link from the first level. Next, checking on the selected species in the panel will open a new browser window with a detailed view of the corresponding allosteric protein being displayed, including sequence, structure, native mutation, protein modification and allosteric description. As with most biological databases, all of the proteins illustrated in ASD are hyperlinked to other online databases or tables like UniProt, GenBank, Enzyme Nomenclature, KEGG or PDB. By hyperlinking to these particular databases, ASD is able to provide considerably more information about allosteric proteins. ‘Index browse’ allows the browsing of any allosteric molecules by their names, which are arranged in alphabetical sequence within each initial letter tab.
In addition to the ‘Browse’ and ‘Search’ options, ASD also offers background glossary and diagram under its ‘Allosteric Wiki’ menu; allosteric news, meeting, references and significant finding under its ‘Hot Topics’ menu; release note, statistical information, ‘Expert’ platform for communication of allosteric information to external experts and data download under its ‘About’ menu; and miscellaneous links to other databases under its ‘Links’ menu.
DISCUSSION AND CONCLUSIONS
ASD is a manually curated database dedicated to allosteries involving receptors and ligands. It is the first online resource of this kind and the data in ASD are freely available to all potential users. We harvested experimentally verified allosteric proteins and modulators from scientific articles. Of 336 allosteric proteins in ASD, 205 (66.96%) proteins catalyze diverse biological reaction as enzyme included by Enzyme Nomenclature (18) and 283 (84.23%) proteins are contained in KEGG (19). Contrary to the proteins, >95% allosteric modulators are not covered by two important bio-active small molecule databases viz, Drugbank (4.49%) (24) and ChEMBL (0.2%) (31). These allosteric data are accompanied by additional information about their structures, functions, related diseases, external links and associated tools, which may provide a valuable resource for research on the allosteric field.
Our initial collection mainly focused on allosteric proteins and their modulators, which have been extensively studied for >50 years. More than 300 proteins were found as allostery and thus allosteric protein is interchangeable with the term allosteric macromolecules. However, the other type of macromolecules, namely the nucleic acids (DNA and RNA), was recently found to match the rule of allostery in several cases (32–34). Considering the fact that these allosteries are not widely confirmed, nucleic acids are not included in the current version but will be added into ASD in the next version.
ASD provides users with both chemical and biological tools for information mining on allosteric molecules. Chemical searches can suggest similar structures of molecules/scaffolds from known allosteric modulators. Such parallel molecules with known targets in ASD can assist in the recognition of potent allosteric targets for query compound, and such comparable scaffolds can facilitate the structural modifications necessary for novel allosteric drug design. On the biological side, there is a tool that supports search based on sequence similarities of proteins. Combining the information in ASD with sequence alignment should allow for the prediction of allostery for proteins with unknown properties. In addition, high homology between the subtypes of family proteins to some extent hampered the progress of functional identification in the field of both chemical biology and drug discovery. The allosteric site of one subtype aligned to other members of a protein family could provide clues to its potential specific features and eventually make it ideal target for experimental validation.
FUTURE DIRECTIONS
As a resource to study the allosteric world, we will continue updating the database with allosteric proteins and their modulators every 6 months and respond to ‘Expert’ request within 1 week. We have also extended allosteric collection and analysis to another type of macromolecule, the nucleic acids including DNA and RNA. On the other hand, in order to clarify the biological behavior of allosteric modulators identified in screenings, a novel algorithm for recognition of allosteric sites is currently being developed. We believe such integrative allosteric data and tools will give users the expanded knowledge needed for the biological and chemical interpretation of each allosteric event.
AVAILABILITY AND REQUIREMENTS
ASD is freely available at http://mdl.shsmu.edu.cn/ASD/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Ministry of Science and Technology (NO2009CB918404, in part); National Basic Research Program of China (973 Program) (2011CB504001, in part); National Natural Science Foundation of China (90813034, 90919021, in part); Science and Technology Commission of Shanghai (08JC1413400, in part); Shanghai PuJiang Program (10PJ406800, 10PJD010, in part); Innovation Program of Shanghai Municipal Education Commission (grant number 09ZZ23, in part).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Dr Xuefeng Lu and Mr Mars for lasting help in setting up the web server and database configuration. We also thank Dr Hanyi Zhuang for testing the database web interface and offering valuable comments.
REFERENCES
- 1.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 2.Lindsley JE, Rutter J. Whence cometh the allosterome? Proc. Natl Acad. Sci. USA. 2006;103:10533–10535. doi: 10.1073/pnas.0604452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 4.Taly A, Corringer PJ, Guedin D, Lestage P, Changeux JP. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat. Rev. Drug Discov. 2009;8:733–750. doi: 10.1038/nrd2927. [DOI] [PubMed] [Google Scholar]
- 5.Munro G, Ahring PK, Mirza NR. Developing analgesics by enhancing spinal inhibition after injury: GABAA receptor subtypes as novel targets. Trends Pharmacol. Sci. 2009;30:453–459. doi: 10.1016/j.tips.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Matschinsky FM. Assessing the potential of glucokinase activators in diabetes therapy. Nat. Rev. Drug. Discov. 2009;8:399–416. doi: 10.1038/nrd2850. [DOI] [PubMed] [Google Scholar]
- 7.Hill AV. The possible effects of the aggregation of the molecules of hæmoglobin on its dissociation curves. J. Physiol. 1910;40:iv–vii. [Google Scholar]
- 8.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J. Mol. Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 9.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 10.Kurganov BI, Iakovlev VA. Allosteric Enzymes: Kinetic Behaviour (English version) NY, USA: John Wiley & Sons Inc.; 1982. [Google Scholar]
- 11.Kamata K, Mitsuya M, Nishimura T, Eiki J, Nagata Y. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 2004;12:429–438. doi: 10.1016/j.str.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villaverde A. Allosteric enzymes as biosensors for molecular diagnosis. FEBS Lett. 2003;554:169–172. doi: 10.1016/s0014-5793(03)01160-8. [DOI] [PubMed] [Google Scholar]
- 14.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogoyevitch MA, Fairlie DP. A new paradigm for protein kinase inhibition: blocking phosphorylation without directly targeting ATP binding. Drug Discov. Today. 2007;12:622–633. doi: 10.1016/j.drudis.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 16.The Universal Protein Resource (UniProt) Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwin CW NC-IUBMB. Enzyme Nomenclature 1992. San Diego, CA, USA: Academic Press; 1992. [Google Scholar]
- 19.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 22.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited–architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, et al. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;36:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 27.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weininger D. SMILES 1. Introduction and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–38. [Google Scholar]
- 31.Overington J. ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J. Comput. Aided Mol. Des. 2009;23:195–198. doi: 10.1007/s10822-009-9260-9. [DOI] [PubMed] [Google Scholar]
- 32.Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc. Natl Acad. Sci. USA. 2009;106:13175–13179. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar D, An CI, Yokobayashi Y. Conditional RNA interference mediated by allosteric ribozyme. J. Am. Chem. Soc. 2009;131:13906–13907. doi: 10.1021/ja905596t. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Ellington AD. Design principles for ligand-sensing, conformation-switching ribozymes. PLoS Comput. Biol. 5:e1000620. doi: 10.1371/journal.pcbi.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]