Abstract
P2CS (http://www.p2cs.org) is a specialized database for prokaryotic two-component systems (TCSs), virtually ubiquitous signalling proteins which regulate a wide range of physiological processes. The primary aim of the database is to annotate and classify TCS proteins from completely sequenced prokaryotic genomes and metagenomes. Information within P2CS can be accessed through a variety of routes—TCS complements can be browsed by metagenome, replicon or sequence cluster (and these genesets are available for download by users). Alternatively a variety of database-wide or taxon-specific searches are supported. Each TCS protein is fully annotated with sequence-feature information including replicon context, while properties of the predicted proteins can be queried against several external prediction servers to suggest homologues, interaction networks, sub-cellular localization and domain complements. Another unique feature of P2CS is the analysis of ORFeomes to identify TCS genes missed during genome annotation. Recent innovations for P2CS include a CGView representation of the distribution of TCS genes around a replicon, categorization of TCS genes based on gene organization, an expanded domain-based classification scheme, a P2CS ‘gene cart’ and categorization on the basis of sequence clusters.
INTRODUCTION
As the number of publicly available prokaryotic genomic and metagenomic data sets escalates, there is an increasing need to catalogue the genes of these data sets in a publically accessible and user-friendly format. Databases of enzymes such as KEGG and BRENDA successfully link genomic data with metabolism (1,2), but particular problems are encountered when dealing with signal-transduction proteins that are typically composed of multiple protein domains. In addition, sequence homologues of signalling proteins often regulate fundamentally different physiological processes, requiring classification by alternative criteria to sequence similarity, such as domain composition (3).
Two-component systems (TCSs) are the dominant prokaryotic-signalling pathway. The average Bacterial genome encodes more than 50 TCS proteins, with significant numbers also found in Archaeal genomes (4). A typical TCS comprises a pair of signal-transduction proteins—a histidine kinase (HK) and partner response regulator (RR)—both of which are multi-domain proteins. HKs usually contain transmembrane sensory (input) domains which upon stimulation activate autophosphorylation of a histidine residue in the HK-transmitter domain. The phosphorylated-transmitter domain then binds to the receiver domain of the partner RR and this leads to transfer of the phosphoryl group to an aspartate residue of the receiver domain. In the majority of cases, phosphorylation of the RR changes the activity of an effector (output) domain within the RR, which brings about a physiological change, often through altered expression of specific genes.
In 2008 the ‘Prokaryotic 2-Component System’ database (P2CS) was made available to the public at http://www.p2cs.org. P2CS originally catalogued the TCS genes of 755 genomes and 39 metagenomes (5). P2CS has been updated recently to include data from 1125 genomes and enriched with several new features including automatic updates, different approaches for the classification of TCS proteins, consideration of gene organization, expanded annotation, a ‘gene cart’ and improvements to the web interface. Compared to more generalized databases of signal-transduction proteins such as MiST, Genome Atlas and SENTRA (6,7,8), P2CS provides a relative diversity of information for each protein entry and a variety of approaches for acquiring and outputting those data.
This is the first description of P2CS in Nucleic Acids Research, and we therefore provide an account of P2CS functionalities, however as P2CS is an established database this report also concentrates on recent upgrades to the database.
P2CS DATABASE
The P2CS database has a modular structure which begins with the importing of genomic and metagenomic data from the NCBI (http://www.ncbi.nlm.nih.gov/) and the IMG/M (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi), respectively, as described previously (5). TCS proteins are identified from protein genomic files, while DNA-sequence data is also scanned and translated to constitute an ORFeome, which is then searched for the presence of mis-predicted TCS genes (those missed by genome annotation). Identification of TCS proteins is achieved through domain analysis of each predicted protein/ORF, assessing the presence of any member of a manually curated list of TCS domains from the SMART and Pfam libraries (5). Identified TCS proteins are then categorized into classes based on domain composition, for example the presence of a receiver domain and the absence of a transmitter domain leads to classification of the protein as a RR. At this stage proteins are classified as RR, HK or phosphotransfer proteins (proteins that are comprised of just an Hpt or HisKA domain, and which can shuttle phosphoryl groups between two receiver domains).
HKs are then sub-classified as classic, hybrid (also containing receiver domains), unorthodox (hybrid HKs also possessing an Hpt phosphotransfer domain), CheA (HKs whose transmitter domain contains an integral Hpt domain, resembling the HK of Escherichia coli chemotaxis CheA) or ‘possible incomplete’ (containing transmitters lacking an obvious phosphorylatable His residue). RRs are sub-classified according to the nature of any output domain present on the protein (5), for instance any RRs containing a Trans_reg_C domain are classified as members of the OmpR family.
Information within P2CS can be accessed through a variety of routes—TCS complements can be browsed by metagenome or replicon (and these genesets are available for download by users). Alternatively, a variety of database-wide or taxon-specific searches are supported. Each TCS protein is fully annotated with sequence-feature information including replicon context, presence of transmembrane helices, domain description and coding bias among others. Summary gene information (gene organization and domain architecture) is also provided in lists accessed by browsing or searching. Properties of the predicted proteins can also be queried against several external prediction servers to suggest homologues, interaction networks, sub-cellular localization and domain complements (Supplementary Figure S1).
NEW FEATURES
Expanded classification
The growing number of sequenced genomes and metagenomes and the increasing diversity of TCS domain combinations that results from this, requires a periodic reappraisal of new domain architectures. This has been accomplished recently for RRs by Galperin (3), and we have implemented this new classification scheme in P2CS. Thus RRs are now divided into 35 families, rather than the original 24. Half of these defined families contain DNA-binding output domains and a third of them regulate their targets using enzyme output domains (Supplementary Figure S2). However, 11% of RRs still cannot be unambiguously assigned to a family on the basis of current criteria, and as further research allows classification schemes to expand to recognize novel families of RRs, P2CS will continue to reflect those changes in knowledge.
Expanded annotation
The pages for individual TCS proteins have been enriched with further annotation and external links (Supplementary Figure S1). Secondary structure plots are available and were computed using the PSIPRED method (9). The result of the protein structure prediction is presented as a summary of the number of strands and helices and their location on the protein sequence (Supplementary Figure S3). In addition further links are provided, allowing the protein to be queried against various prediction servers. New links allow navigation to Genbank entries for TCS proteins, prediction of interaction networks through STRING (10), assessment of likely sub-cellular compartmentalization through a variety of ‘Signal Search’ links, to Psortb, Phobius, LipoP and SignalP (11–14). Detailed domain descriptions can also be retrieved using links to servers such as CDD, SMART and Pfam (15–17).
TCS clustering
In addition to classifying TCS proteins on the basis of domain complement, an alternative approach has been devised based on clustering proteins by sequence similarity (70% identity). The CD-HIT algorithm (18) was used to cluster all the TCS proteins in P2CS, and exploration of the different clusters can be achieved through a new ‘Clusters’ submenu within the Browse menu, alongside ‘Genomes’ and ‘Metagenomes’ (Figure 1). Alternatively, the clustering information is displayed for all the TCS proteins through each protein page (Supplementary Figure S1).
Figure 1.
A submenu displaying details of membership of a particular TCS-gene cluster. There are several sequence members of this cluster defined by P2CS annotation, in addition to members with solved structures.
Included within the clustering were structure sequences from the PDB database, so that when a cluster contained a protein of known 3D structure (and in some cases several), a prediction of 3D structure could be made for all TCS proteins in the cluster (239 clusters contained a member with solved structure), and a link established between these proteins and PubMed literature (Figure 1). The cluster containing the largest number of sequences is cluster number 5475 with 151 sequences and four PDB structures. As expected, it includes members of the most abundant RR family (OmpR). Cluster 8225 is characterized by a considerable number of 3D structures (87) and contains essentially CheY family RRs.
Clustering data can be utilized by biologists in a variety of ways, and we provide an example here. The RR BL02604 of Bacillus licheniformis ATCC 14580 (GI:52081663) is encoded adjacent to a mis-predicted HK. Accessing the clustering data for BL02604 shows three other RR genes in the cluster, each of which is adjacent to a HK. Such information provides added weight to the identification of the mis-predicted HK, suggesting that BL02604 is part of a pair rather than an orphan—and immediately suggests the identity of the partner HK.
Genetic organization
Another recent innovation for P2CS is the analysis of genetic organization. Many TCS genes are encoded in pairs, or in more complex arrangements, and gene organization can provide insights into signalling partnerships. P2CS implements a categorization scheme whereby TCS genes are considered to be ‘linked’ within a locus if within 200 bp of each other and encoded on the same strand. TCS gene loci are categorized as orphans (one gene), pairs (two genes), triads (three genes), tetrads (four genes) or pentads+ (five or more genes). In the current version of P2CS-containing 77357 TCS genes, triads and tetrads are relatively common (5% of all TCS genes). However, larger clusters are rare (26 pentads, five hexads, one heptad and one decad, together accounting for only 0.2% of all TCS genes). The decad is a particularly intriguing locus comprising genes Glov_1564 to Glov_1573 from Geobacter lovleyi SZ, together encoding nine receiver and three transmitter domains. Gene organization summaries are presented on each replicon page, while each gene page describes the gene organization and allows navigation to other genes in the locus.
Web interface improvements
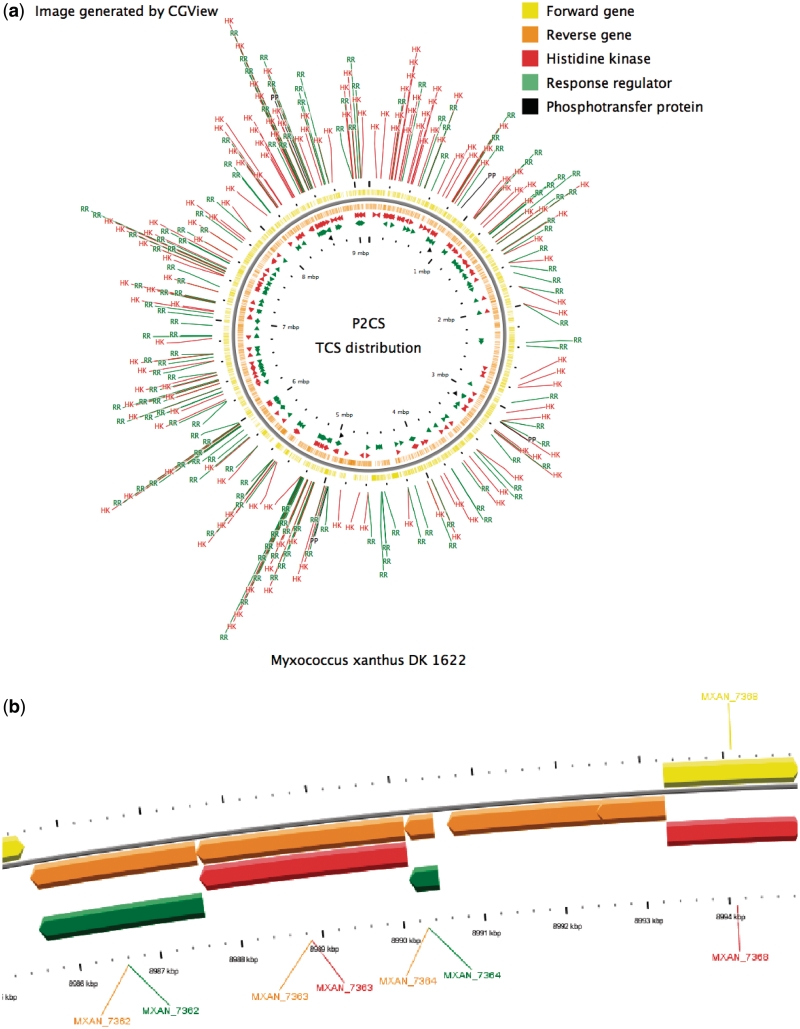
Other significant improvements to the P2CS user interface have also been implemented. Each replicon summary page (Supplementary Figure S4) now provides a depiction of the distribution of TCS genes around the replicon generated using CGView (19). This image can be enlarged (Figure 2a), or opened as a zoomable map (Figure 2b) and supplements the information obtained regarding genetic organization. A sequence logo of the region around the phosphorylatable HK histidine residue (H-box) is also shown for each replicon. Using the WebLogo tool (20), we generated a graphical representation of the pattern of sequence conservation of all the predicted HKs belonging to each replicon (Supplementary Figure S4). This is the sequence region used to identify putative phosphorylation sites in possible incomplete HKs (5).
Figure 2.
CGView representation of the distribution of TCS genes around a replicon (Myxococcus xanthus shown as an example) (a) and a close-up of a TCS gene locus (b).
P2CS cart
A shopping-cart function has been added to P2CS that can be populated by genes individually, or en masse from gene lists. The P2CS cart (Supplementary Figure S5) can then be accessed through the browse menu. As with other sets of TCS genes, the cart is downloadable in a variety of formats (Excel, nucleotide FASTA, amino acid FASTA or tab-delimited).
Automatic update
Previously, P2CS was updated with new genomes yearly, however since May 2010 the database has been upgraded to update automatically every 2 months. The suite of programs is designed to look for new genomes, launch the analysis system and update itself automatically. The results are then immediately visible and consultable on the user interface.
DISCUSSION AND FUTURE DIRECTIONS
P2CS provides a one-stop shop for analysis of the TCS encoded by completely sequenced genomes and metagenomes. The site is designed for use by experimental biologists rather than bioinformaticians and recent innovations have made the user interface even more intuitive and accessible. The P2CS site is distinct from other public signal transduction databases because it contains computational analysis of the modular TCSs of prokaryotic genomes and metagenomes and provides a complete overview of information on each TCS and the TCSs within each replicon, including predicted candidate proteins and probable proteins, which need further curation/validation.
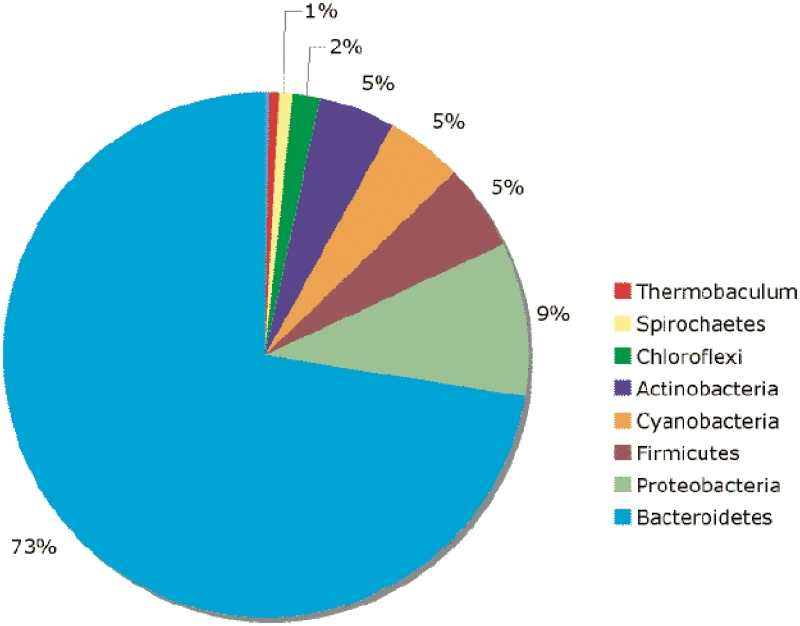
Because of the flexible ways in which data can be retrieved from P2CS—through a variety of searches, or by browsing—it is increasingly facile to perform sophisticated analyses on the genomics of TCS without requiring any specialist scripting or programming skills. For example, simple searches based on domain architecture highlight an unusual group of 249 hybrid kinases that each contains a C-terminal output domain (Supplementary Table S1). The majority of these proteins (73%) come from Bacteroidete genomes (Figure 3), with 40 alone from Bacteroides thetaiotaomicron VPI-5482 (Supplementary Table S1). The output domain of these unusual hybrid kinases is an AraC-like DNA binding domain in most cases (72%). The prevalence of hybrid kinases in Bacteroidete genomes has been noted before (21) as has the prevalence of HTH_AraC output domains in Bacteroidete TCSs (22). However, relaxing the requirement that an output domain be C-terminal another 52 unusual hybrid kinases are uncovered (Supplementary Table S2), the majority of which have the domain geometry Receiver, SpoIIE, Transmitter (with one example where the SpoIIE domain is replaced by a Trans_Reg_C domain) and which are found exclusively in Proteobacterial genomes.
Figure 3.
Taxonomic distribution of unusual hybrid HKs with C-terminal output domains.
Above we have described novel developments of the P2CS database and the unique features it provides. Recent innovations for P2CS include automatic 2-monthly update, a revised domain-based categorization scheme, novel cluster-based classification, together with analysis of gene organizations. In the future, we will continue our efforts to add predictions for new genomes and integrate more metagenomes. Currently, P2CS establishes a link between 3D structures, the clustering module and PubMed literature. We plan to expand this facility to provide more information within the database. Subsequent versions of our system will also incorporate phylogenomic analyses based on the TULIP tool (23), multiple alignments visualization and clustering methods will take into account more criteria. The development of P2CS is user-guided, for instance the choice to open certain pages in new windows was made in response to user requests. We continue to encourage comments from users and respond to developments in the community, such as implementing expanded classification schemes as they become available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: UMR 6191 - CEA, CNRS, Aix-Marseille Université.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to DSV/IBITEC-S/GIPSI team and particularly Arnaud Martel and Jean-Marc Le Failler for the hosting server installation.
REFERENCES
- 1.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galperin MY. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010;13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wuichet K, Cantwell BJ, Zhulin IB. Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 2010;13:219–225. doi: 10.1016/j.mib.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barakat M, Ortet P, Jourlin-Castelli C, Ansaldi M, Méjean V, Whitworth DE. P2CS: a two-component system resource for prokaryotic signal transduction research. BMC Genomics. 2009;10:315. doi: 10.1186/1471-2164-10-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2009;38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiil K, Ferchaud JB, David C, Binnewies TT, Wu H, Sicheritz-Ponten T, Willenbrock H, Ussery DW. Genome update: distribution of two-component transduction systems in 250 bacterial genomes. Microbiol. 2005;151:3447–3452. doi: 10.1099/mic.0.28423-0. [DOI] [PubMed] [Google Scholar]
- 8.D’Souza M, Glass EM, Syed MH, Zhang Y, Rodriguez A, Maltsev N, Galperin MY. Sentra: a database of signal transduction proteins for comparative genome analysis. Nucleic Acids Res. 2007;35:D271–D273. doi: 10.1093/nar/gkl949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 10.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 19.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 20.Crooks GE, Hon G, Chandonia J, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghavan V, Groisman EA. Orphan and hybrid two-component system proteins in health and disease. Curr. Opin. Microbiol. 2010;13:226–231. doi: 10.1016/j.mib.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitworth DE, Cock PJA. Two-component systems of the myxobacteria: structure, diversity and evolutionary relationships. Microbiology. 2008;154:360–372. doi: 10.1099/mic.0.2007/013672-0. [DOI] [PubMed] [Google Scholar]
- 23.Bastien O, Ortet P, Roy S, Maréchal E. A configuration space of homologous proteins conserving mutual information and allowing a phylogeny inference based on pair-wise Z-score probabilities. BMC Bioinformatics. 2005;6:49. doi: 10.1186/1471-2105-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]