Abstract
Indians, representing about one-sixth of the world population, consist of several thousands of endogamous groups with strong potential for excess of recessive diseases. However, no database is available on Indian population with comprehensive information on the diseases common in the country. To address this issue, we present Indian Genetic Disease Database (IGDD) release 1.0 (http://www.igdd.iicb.res.in)—an integrated and curated repository of growing number of mutation data on common genetic diseases afflicting the Indian populations. Currently the database covers 52 diseases with information on 5760 individuals carrying the mutant alleles of causal genes. Information on locus heterogeneity, type of mutation, clinical and biochemical data, geographical location and common mutations are furnished based on published literature. The database is currently designed to work best with Internet Explorer 8 (optimal resolution 1440 × 900) and it can be searched based on disease of interest, causal gene, type of mutation and geographical location of the patients or carriers. Provisions have been made for deposition of new data and logistics for regular updation of the database. The IGDD web portal, planned to be made freely available, contains user-friendly interfaces and is expected to be highly useful to the geneticists, clinicians, biologists and patient support groups of various genetic diseases.
INTRODUCTION
The load of genetic diseases varies widely between different populations depending on its structure, reproductive practices and other factors. Control and management of the genetic disorders depend on identification of the variants in the genome that are causally linked with the disease. The spectrum of such variants, i.e. mutations, is different in different population groups. Remarkable progress has been made towards capturing the genomic variation in the context of genetic diseases with the advancement of DNA sequencing technologies, the capacity to handle large amount of data by building databases and faster dissemination of information through the worldwide web. It is, therefore, not surprising that the initial modest beginning of Mendelian Inheritance of Man (MIM) transformed later to Online MIM (OMIM). Currently, the most expanded version of database specifically cataloging the mutations relating genetic diseases across globe is Human Gene Mutation Database (HGMD). In addition, special interest groups generated ‘locus specific databases’ (LSDBs) and lately ‘national and ethnic mutation databases’ (NEMDBs) have also emerged containing mutational data for specific countries (Table 1). Such endeavor enormously boosts the efforts related to diagnosis of genetic diseases, detection of carriers for disease management and control and genetic counseling to mitigate the suffering of the affected families. However, no such database on genetic diseases exists for India, a country inhabited by more than a billion people and predicted to have a high load of recessive disorders in the population.
Table 1.
IGDD compared to existinga NEMDBs (National and Ethnic Mutation Databases)
| Databases | Country population (in millions) | Patients/ carriers studied | Diseases | Total mutations recorded | Unique mutations | Patient- specific records | Summary statistics provided | Launched/ last updated | Published |
|---|---|---|---|---|---|---|---|---|---|
| Finnish Disease Database (Finland) | 5.30 | INR | 35 | 1362b | INR | No | No | 2002 | Yes (1) |
| Iranian Human Mutation Database (Iran) | 68.69 | INR | 98 | 466 | 415 | No | Yes | September 2003 | No |
| The Cypriot National Mutation Frequency Database (Cyprus) | 1.05 | INR | 19 | 1478 | 85 | No | No | August 2006 | Yes (2) |
| The Hellenic National Mutation Database (Greece) | 10.68 | INR | 14 | 3179 | 221 | No | No | June 2006 | Yes (3) |
| The Iranian National Mutation Frequency Database (Iran) | 68.69 | INR | 8 | 2614 | 74 | No | No | August 2006 | Yes (2) |
| The Israeli National Genetic Database (Israel) | 7.60 | INR | 330 | 2581 | 904 | No | No | July 2010 | Yes (4) |
| The Lebanese National Mutation Frequency Database (Lebanon) | 0.02 | INR | 6 | 880 | 60 | No | No | January 2006 | Yes (5) |
| The Moroccan Human Mutation Database (Morocco) | 28.56 | INR | 138 | INR | 229 | No | No | February 2010 | Yes (6) |
| The Serbian National Mutation Frequency Database (Serbia) | 7.78 | INR | 6 | 68c | 68 | No | No | April 2006 | No |
| Thailand Human Mutation and Variation database (Thailand) | 66.40 | INR | 119 | 589 | 518 | No | Yes | August 2008 | Yes (7) |
| Turkish Human Mutation Database (Turkey) | 71.51 | INR | 2 | 57c | 57 | No | No | January2006 | No |
| FINDbase worldwide (92 populations) | NA | INR | 32 | 3553 | 1226 | No | Yes | June 2009 | Yes (8) |
| Indian Genetic Disease Database (India) | 1180.16 | 5760 | 52 | 6647 | 780 | Yesd | Yes | August 2010 | This report |
NA: Not applicable; INR: Information not retrievable.
aCurrently available/accessible online; Singapore Human Mutation and Polymorphism Database is not included since the variants listed in the database are not distinctly categorized into ‘mutations’ or ‘polymorphisms’.
bNot specified whether total or unique mutations.
cDatabase records only unique mutations.
dPatient-specific record of IGDD includes personal data (e.g. age, sex, ethnicity, geographical location, etc.) and clinical and bio-chemical data.
The evolutionary history of primitive Indian ethnic groups and migration from Africa, middle-east and west Asia, southern China and south-east Asia has added to the genetic diversity of the country (9). However, religion, language and geographical location of habitat serve as barriers to random mating in the Indian population. Inbreeding is practiced in some geographical regions of India (population-inbreeding coefficient: 0.00 to 0.20) (10). Thus, the overall heterogeneity of population along with the underlying endogamy makes India, a unique case of importance with respect to a high prevalence of genetic diseases and mutations. This highlights the importance of identifying recessive diseases in the Indian groups and screening the causal genes. In addition to the overall effect of ‘founder events’, in some communities, the load of genetic disorder is relatively higher due to the practice of consanguineous marriage, especially in south India (11).
In March 2006, a study conducted through the March of Dimes Birth Defect Foundation, reported the birth defect prevalence in India as 64.4 (per 1000 live births) (12). Rao and Ghosh (2005) report that 1 out of 20 children admitted to hospital has a genetic disorder that ultimately account for about 1 out of 10 childhood deaths (13). In India’s urban areas, congenital malformations and genetic disorders are the third most common cause of mortality in newborns (14). However, there is no common source of information to assess the load of specific genetic diseases reported in India, extent of locus and mutational heterogeneity, common mutations in the causal genes and the extent of molecular studies carried out vis-à-vis lack of it in the context of the disease load. In fact, most of the pilot studies are local and hospital based. The genetic services are also not well established and localized sporadically. The situation certainly calls for a comprehensive repository of mutational data aided by specific clinical and other relevant information of patients from different regions of India. Here we describe Indian Genetic Disease Database (IGDD), a comprehensive documentation that intends to record patient-specific mutation spectrum of genetic diseases among the Indian population that would help designing assays and diagnostic tests to detect mutations, diagnose genetic diseases and identify carriers.
DATABASE ORGANIZATION
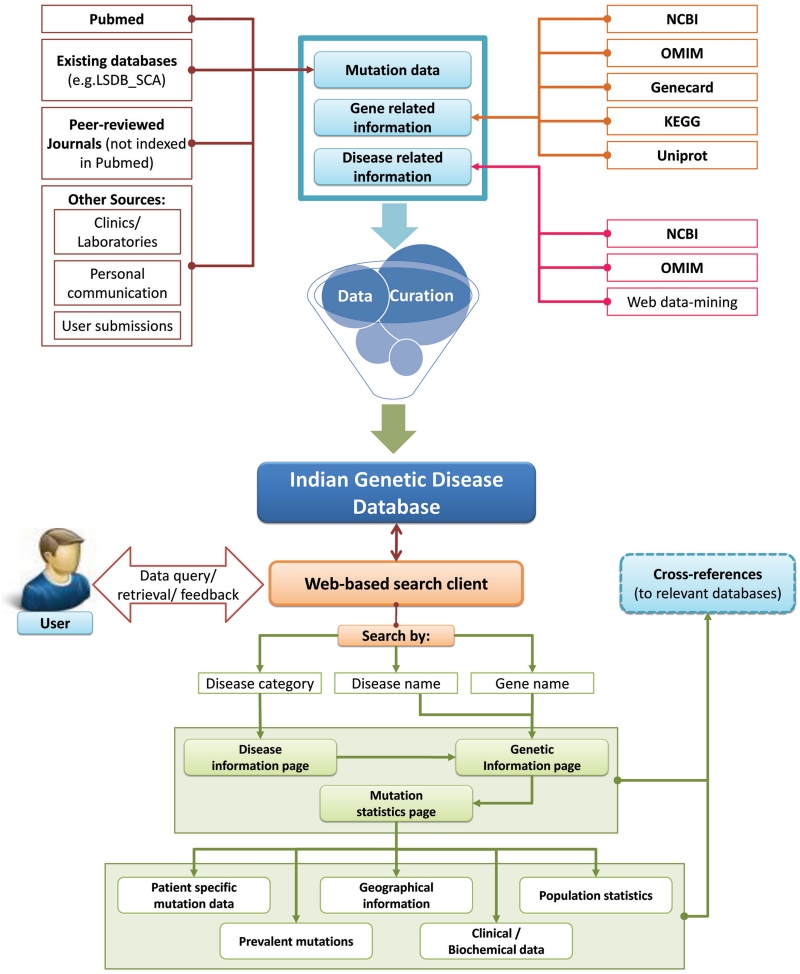
The logistics based on which IGDD has been created is shown schematically in Figure 1. The database offers an integrated and curated repository of experimentally characterized and reported mutations responsible for genetic disorders in Indian population. An easy-to-use web interface allows a remote user to retrieve (and submit) data through interactive web forms. The home page of IGDD provides links to other major public-domain knowledge-bases on human genetic disorders. Details of the software design, data sources, query options and other features of the database are described in the following subsections.
Figure 1.
The schematic representation of the IGDD.
Software design and implementation
The database is designed and implemented on a three-tier architecture—user/client, web-interface and RDBMS backend. The web interface is comprised of a collection of ‘web applications’/‘web forms’ developed in Microsoft Visual Basic .NET 2003. The home page of the database (http://www.igdd.iicb.res.in) serves as the gateway to the interlinked web forms capable of querying the database contents dynamically as instructed (by the user) through button clicks, check-boxes and drop-down menus. In the backend, the relational database is managed with ORACLE 9i. The data collected from different sources are initially stored in manually curated flat-files and uploaded to the database through the SQL*Loader utility. Statistics and figures accompanying the data are auto-generated by software tools developed in-house and subject to automated revision during each update. The database is currently designed to work best with Microsoft Internet Explorer 8 (optimal resolution 1440 × 900).
Source of data
The primary source of data is peer-reviewed published reports. With exception of a few reports all others are cited in PUBMED. In addition, data have been collected through personal communication with genetic laboratories, especially in case of β-thalassemia—the most prevalent genetic disease in India. All the data sources are duly referred to and respective bibliographic pages are hyperlinked.
For convenience of users, the diseases enlisted in IGDD have been divided into various categories such as ‘Blood Related Disorders’, ‘Eye related Disorders’, ‘Pigmentation Disorders’, etc. Diseases with complex clinical syndromes or affecting multiple organs have been included under the ‘Multisystem Disorder’ category. Every documented disorder has been described briefly and aided by proper links (to OMIM) for more detailed reading.
Data content
IGDD release 1.0 holds entries for 52 genetic diseases and 63 related genes collated from 123 reports, published during 1993–2010. Currently, 2394 patients and 3366 carriers (resident or non-resident Indian individuals) are enlisted in the database harboring 6647 mutations of which 780 are unique in nature. Majority of these mutations are missense changes (41.3%) followed by other types of mutations (Table 2).
Table 2.
Summary of the raw data of the IGDD
| Parameters | Counts |
|---|---|
| Patients | 2394 |
| Carriers | 3366 |
| Male | 920 |
| Female | 276 |
| Sex not specified | 4564 |
| Diseases/disorders/syndromes | 52 |
| Disease with known mode of inheritance | 51 |
| Autosomal dominant | 12 |
| Autosomal recessive | 29 |
| X-linked dominant | 1 |
| X-linked recessive | 6 |
| Y-linked | 0 |
| Complex | 1 |
| Multiple | 2 |
| Genes | 63 |
| Total mutations | 6647 |
| Unique mutations | 780 |
| Missense mutations | 322 |
| Nonsense mutations | 70 |
| Deletion mutations | 91 |
| Insertion mutations | 49 |
| InDel mutations | 8 |
| Splice site mutations | 48 |
| Repeat mutations | 85 |
| Gross mutations | 106 |
| Synonymous mutations | 1 |
| Total reports studied | 123 |
| Time span (in years) | 1993–2010 |
Data curation
The errors found in report of mutations have been corrected when it is obvious. Those variants have not been included in the database for which coordinates of the nucleotide in the gene/cDNA and type of mutation are not clearly presented. All the mutations in the database have been linked to specific individuals with their respective phenotypic data depending on the availability of such information. Those studies that reported total mutations only, without any patient record or the number of alleles, were not enlisted in the database. Attempts are being made to convert all the mutations in single format as recommended by the Human Genome Variation Society (HGVS).
Query options
IGDD can be navigated through by three major query options: (i) disease category, (i) disease name and (iii) gene name, as depicted in Figure 1. Selection for a specific disease category through respective buttons directs the users to the ‘Disease Information’ page, displaying the list of diseases under the preselected category, along with short description. Selection of a specific disease, either through the buttons in the Disease information page, or directly from a drop-down menu provided in the search bar routes the users to a 'Genetic Information' page that lists the causal genes, their chromosomal locations and subtypes of the disease, wherever relevant. This page may also be accessed by selecting the respective gene from a drop-down menu in the search bar. Each of the enlisted genes is linked to a ‘Mutation Statistics’ page that displays information on the encoded protein and mutation statistics along with cross references to global databases, LSDBs and Disease-Support groups.
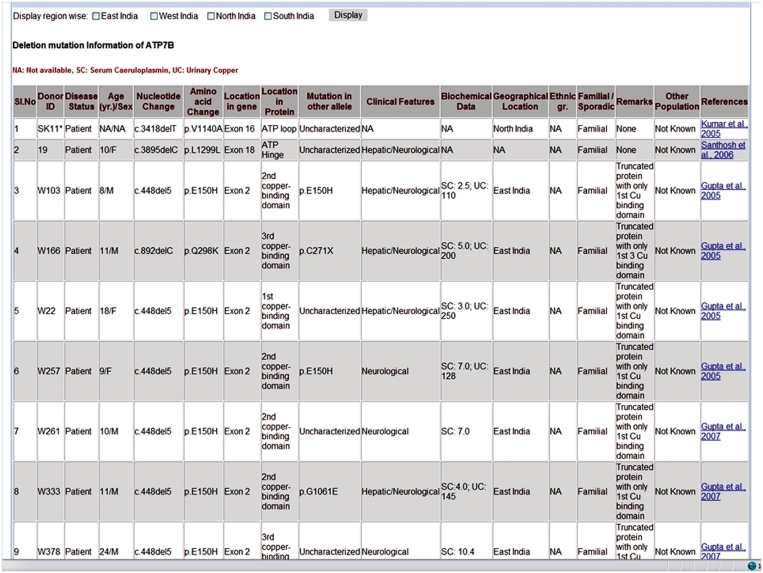
A second level of query options is provided in the Mutation Statistics page through which the users can select for a specific type of mutation to arrive at the respective Mutation page. Figure 2 shows a screen shot of the ‘Mutation page’ that displays available individual specific-information. A search tool has been incorporated in this page to allow the user to search the relevant data for a specific mutation, either by nucleotide change or amino acid change. Moreover, a filtering utility helps the user identify mutations reported from different geographical locations of India.
Figure 2.
A screen-shot of the Mutation Page.
The prevalent mutations for each disease gene (where n > 50) are graphically represented in the ‘Mutation Statistics’ page. The number of individuals harboring the mutations pertaining to a specific disease from different geographical locations is pictorially represented in the Indian map. To make best of data accessibility, the summary statistics for each disease gene has been provided as a downloadable text file (Summary sheet) in the Mutation Statistics page. A detailed users’ manual is available in the ‘Help Page’ to facilitate effective usage of the database.
Data submission and updates
There is a provision for submission of new mutation data in the database. We shall accept both novel and previously reported mutations identified in new patients that would help project the mutational load in different population groups in India. Currently, mutation submission can be done by sending a duly filled submission form and sent via email (igdd.iicb@gmail.com). However, mutational data will be accepted based on either their publication in peer-reviewed journal or supportive documentary evidence leading to identification of the mutations. We plan to make the submission a web-based feature in near future for user convenience. All updates would be incorporated in the updated versions of the database planned to be released every 4–6 months interval depending on the volume of new data available.
DATABASE AVAILABILITY
The database would be publicly available free of cost without any license fees or requirement of prior registration.
SALIENT FEATURES OF THE DATABASE
At present, IGDD represents one of the most data-intensive repositories compared to other available NEMDBs (Table 1). It can be used as a platform to analyze and retrieve maximum information on disease prevalence trend, common mutations and most importantly the clinico-pathological data associated with specific mutations for a particular genetic disorder. In this context, unlike most other mutation databases, IGDD has been formatted as individual centric to correlate the genotype of an individual with his/her disease-related phenotype. Thus genotype–phenotype correlation could be attempted and compared between different individuals (i) who are homozygous for the same mutation or (ii) bear different mutations with similar fate of the encoded protein (e.g. different termination mutations, gross deletion, etc.). Further enrichment of the database for this purpose would depend on the input from the investigators and we plan to make an effort toward this goal. However, since >74% of Indians inhabit in the rural areas with limited medical care and accessibility to diagnostic centers, the load of genetic diseases is expected to be much higher than projected through the database.
CONCLUSION
Genetic diseases can be controlled best through an integrative approach of community education, population screening, genetic counseling, carrier identification and neonatal screening. IGDD would provide a key platform for clinicians, epidemiologists, geneticists and genetic counselors to access a central genetic data-source for the Indian population. This centralized mutation database is likely to play a valuable role in correlation of genotype with phenotype. We think that over long time, with enrichment of the database, the benefits accrued from it would apply to other countries (e.g. Pakistan, Bangladesh, Srilanka, Bhutan and Nepal) of the Indian subcontinent that share historically similar population groups divided by political boundaries. In addition, such implication is more directly applicable to the nonresident Indians across the world migrated in relatively recent past.
FUNDING
Council of Scientific and Industrial Research (CSIR), India; Department of Biotechnology (DBT), India (Grant no. BT/BI/04/055-2001); Senior Research Fellowship awards from CSIR, Government of India (to S.P., M.S. and A.D.). Funding for open access charge: CSIR (partial).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ms Shilpee Pal for her efforts in collation of data, Dr P Sundaresan (Aravind Eye Hospital, Madurai), Dr Sila Chakrabarty (Institute of Haematology and Transfusion Medicine, Calcutta Medical College and Hospital, Kolkata), Prof. Uma Dasgupta (Calcutta University) and Prof. Nitai P. Bhattacharyya (Saha Institute of Nuclear Physics, Kolkata) for providing patient records and information for some of the diseases.
REFERENCES
- 1.Sipilä K, Aula P. Database for the mutations of the Finnish disease heritage. Hum. Mutat. 2002;19:16–22. doi: 10.1002/humu.10019. [DOI] [PubMed] [Google Scholar]
- 2.Kleanthous M, Patsalis PC, Drousiotou A, Motazacker M, Christodoulou K, Cariolou M, Baysal E, Khrizi K, Moghimi B, Pourfarzad F, et al. The cypriot and Iranian national mutation frequency databases. Hum. Mutat. 2006;27:598–599. doi: 10.1002/humu.9422. [DOI] [PubMed] [Google Scholar]
- 3.Patrinos GP, van Baal S, Petersen MB, Papadakis MN. Hellenic National Mutation database: a prototype database for mutations leading to inherited disorders in the Hellenic population. Hum. Mutat. 2005;25:327–333. doi: 10.1002/humu.20157. [DOI] [PubMed] [Google Scholar]
- 4.Zlotogora J, van Baal S, Patrinos GP. Documentation of inherited disorders and mutation frequencies in the different religious communities in Israel in the Israeli National Genetic Database. Hum. Mutat. 2007;28:944–949. doi: 10.1002/humu.20551. [DOI] [PubMed] [Google Scholar]
- 5.Megarbane A, Chouery E, van Baal S, Patrinos GP. The Lebanese National Mutation Frequency database. Eur. J. Hum. Genet. 2006;(Suppl. 1):65. [Google Scholar]
- 6.Ratbi I, Gati AE, Sefiani A. The Moroccan human mutation database. Indian J. Hum. Genet. 2008;14:106–107. doi: 10.4103/0971-6866.45004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruangrit U, Srikummool M, Assawamakin A, Ngamphiw C, Chuechote S, Thaiprasarnsup V, Agavatpanitch G, Pasomsab E, Yenchitsomanus PT, Mahasirimongkol S, et al. Thailand mutation and variation database (ThaiMUT) Hum. Mutat. 2008;29:E68–E75. doi: 10.1002/humu.20787. [DOI] [PubMed] [Google Scholar]
- 8.van Baal S, Kaimakis P, Phommarinh M, Koumbi D, Cuppens H, Riccardino F, Macek M, Jr, Scriver CR, Patrinos GP. FINDbase: a relational database recording frequencies of genetic defects leading to inherited disorders worldwide. Nucleic Acids Res. 2007;35:D690–D695. doi: 10.1093/nar/gkl934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadgil M, Shambu Prasad UV, Manoharan S, Patil S, Joshi NV. Peopling of India. In: Balasubramanian D, Appaji Rao N, editors. Hyderabad: The Indian Human Heritage, Universities Press; 1997. pp. 100–129. [Google Scholar]
- 10.Indian Genome Variation Consortium. The Indian Genome Variation database (IGVdb): a project overview. Hum. Genet. 2005;118:1–11. doi: 10.1007/s00439-005-0009-9. [DOI] [PubMed] [Google Scholar]
- 11.Chandrasekhar A, Jayraj JS, Rao PS. Consanguinity and its trend in a Mendelian Population of Andhra Pradesh. Soc. Biol. 1993;40:244–247. doi: 10.1080/19485565.1993.9988850. [DOI] [PubMed] [Google Scholar]
- 12.Christianson A, Howson CP, Modell B. March of Dimes global report on birth defects: the hidden toll of dying and disabled children. March of Dimes Birth Defects Foundation. 2006:33. [Google Scholar]
- 13.Rao VB, Ghosh K. Chromosomal variants and genetic diseases. Int. J. Hum. Gen. 2005;11:59–60. [Google Scholar]
- 14. Identifying regional priorities in the area of human genetics in SEAR: report of an Intercountry Consultation, Bangkok, Thailand, 23–25 September 2003. New Delhi, World Health Organization Regional Office for South-East Asia, 2004 (SEA-RES-121) [Google Scholar]