Abstract
Since its inception in 1994, The RNA Modification Database (RNAMDB, http://rna-mdb.cas.albany.edu/RNAmods/) has served as a focal point for information pertaining to naturally occurring RNA modifications. In its current state, the database employs an easy-to-use, searchable interface for obtaining detailed data on the 109 currently known RNA modifications. Each entry provides the chemical structure, common name and symbol, elemental composition and mass, CA registry numbers and index name, phylogenetic source, type of RNA species in which it is found, and references to the first reported structure determination and synthesis. Though newly transferred in its entirety to The RNA Institute, the RNAMDB continues to grow with two notable additions, agmatidine and 8-methyladenosine, appended in the last year. The RNA Modification Database is staying up-to-date with significant improvements being prepared for inclusion within the next year and the following year. The expanded future role of The RNA Modification Database will be to serve as a primary information portal for researchers across the entire spectrum of RNA-related research.
INTRODUCTION
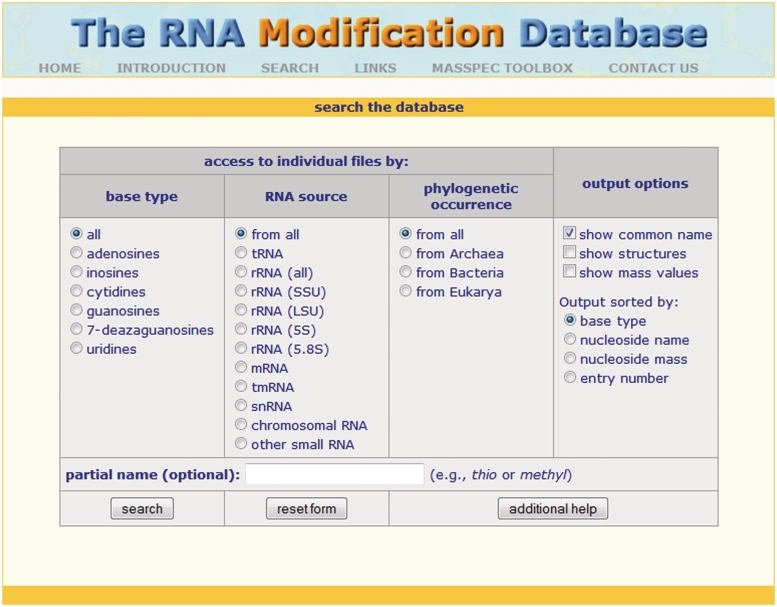
The chemical composition of an RNA molecule allows for its inherent ability to play many roles within biological systems. This ability is further enhanced through the site selected addition of the 109 currently known post-transcriptional modifications catalyzed by specific RNA modification enzymes (1). These naturally-occurring modifications are found in all three major RNA species (tRNA, mRNA and rRNA) in all three primary phylogenetic domains (Archaea, Bacteria and Eukarya) as well as in a handful of other RNA species such as snRNA (2–5) (Table 1). The modifications are one of the most evolutionarily conserved properties of RNAs. Due in large part to comprehensive investigations into the structural and functional roles of modified nucleotides in tRNA, significant advancements have been achieved in our understanding of the various roles played by these modifications (2,6–9). The need to provide a comprehensive, searchable database to house this wealth of knowledge led to the first iteration of The RNA Modification Database (RNAMDB) in 1994 (10). The current version of the database, now housed at The RNA Institute at the University at Albany SUNY, contains all naturally-occurring, RNA-derived modified ribonucleosides for which the chemical structures are known. The RNAMDB provides a user-friendly, searchable interface that directs the user to a detailed information page for each database entry (Figure 1).
Table 1.
The RNA modifications in the database are present in all phylogenetic groups and in many of the known RNA species
| RNA | Phylogenetic source |
||
|---|---|---|---|
| Archaea | Bacteria | Eukarya | |
| tRNA | 43 | 45 | 51 |
| rRNA | 1a | 4a | |
| SSU | 11 | 8 | 18 |
| LSU | 8 | 15 | 12 |
| 5S | 3 | 1 | |
| 5.8S | 5 | ||
| mRNA | 13 | ||
| tmRNA | 2 | ||
| snRNA | 11 | ||
| Chromosomal RNA | 2 | ||
| Other Small RNA | 1 | ||
aDenotes rRNA modifications in which the subunit of origin is either unknown or reported as a mixture of subunits.
Figure 1.
The user-friendly search page makes it easy for users to find RNA modifications in the database based on a variety of different criteria.
Users are invited to submit comments regarding existing entries, including errors and omissions, as well as suggestions for improvements to the following email address: rnamdb@albany.edu.
DATABASE AND CONTENTS
A comprehensive introduction page provides a detailed overview of the current state of the RNAMDB, an in-depth description of the entries and a page specifically dedicated to modifications found in ribosomal RNA. The overview section of the introduction page provides a focus on commonly used symbols representing RNA modifications, reasons for exclusion of certain modified nucleosides and an extensive listing of reviews for further reference.
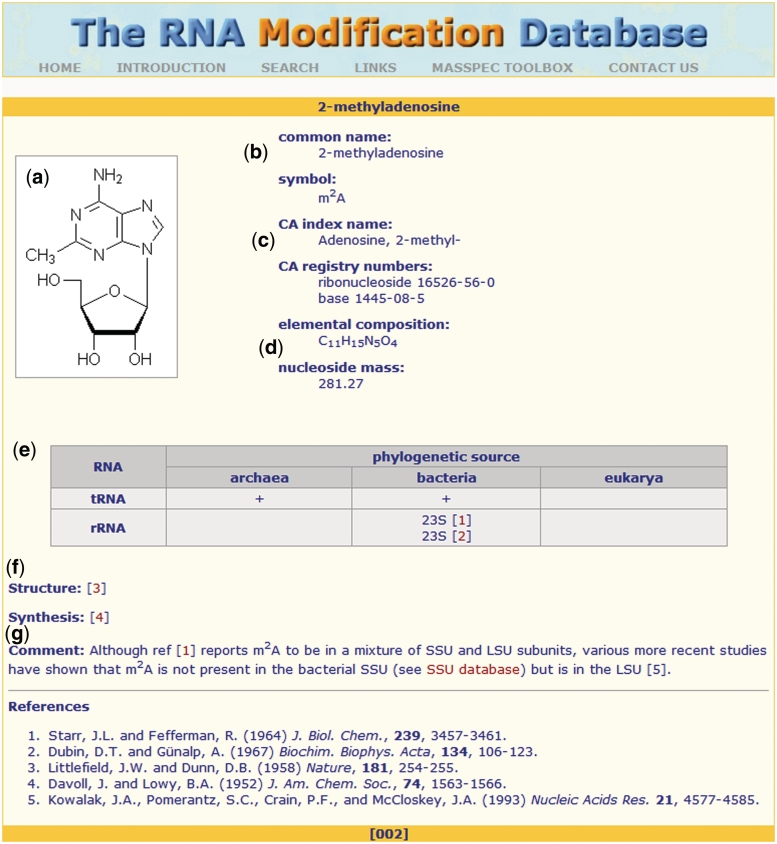
Presently, the information page for each RNAMDB entry contains the following details (Figure 2a–g).
Chemical structure of the modified nucleoside: this structure is simplistic and is not meant to imply the preferred orientation of dihedral angles or sugar pucker.
Common name and symbol.
The searchable Chemical Abstracts registry numbers and index name, which may provide stereochemical information not shown in the chemical structure.
Elemental composition and molecular weight.
Phylogenetic source of the RNA (i.e. Archaea, Bacteria or Eukarya) and the type of RNA species in which it is found (i.e. tRNA, rRNA, etc.).
Literature references for the first reported structure and synthesis. Reports of more recent synthesis or refinements can be found using the Chemical Abstracts registry numbers noted above.
Comments pertaining to any of the information provided on the entry information page provided by database curators.
Figure 2.
The entry information page for 2-methyladenosine shows a typical representation of the statistics that are contained within the database.
An example of a single entry information page for 2-methyladenosine has been provided (Figure 2). The RNAMDB contains 109 such entries that consist of 93 found in tRNAs, 31 in rRNA, 13 in mRNA and 14 in other RNA species such as snRNA, snoRNA and miRNA (3,4,6) (Table 1).
In addition to the Introduction and Search pages, the website contains a compilation of links to other helpful databases and search tools, and provides access to a suite of programs, Masspec Toolbox, designed to assist in the identification and characterization of nucleic acids by mass spectrometry (MS). These dedicated programs are capable of calculating the mass of ribo- and deoxyribo-oligonucleotides from their sequence, as well as predicting the masses of products obtained by submitting these molecules to tandem mass spectrometry (MS/MS), or to digestion by endo- and exonucleases. Other tools can calculate nucleotide and elemental composition from a certain mass and provide the corresponding isotopic distribution. All of these tools allow for users to account for the presence of chemically modified nucleotide residues.
RECENT ADDITIONS
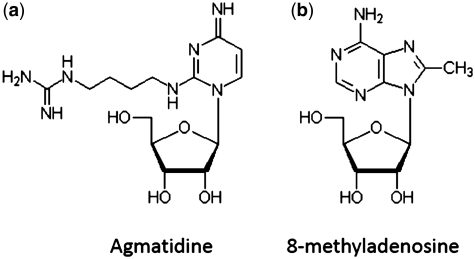
Since the publication of the last database overview (3), 14 new entries have been added to the RNAMDB, including agmatidine and 8-methyladenosine, which have both been added within the past year (11–25) (Table 2). Agmatidine (C+) was recently found in archaeal AUA-decoding tRNAIle2, where it performs a function similar to lysidine, a modified cytidine residue containing lysine in place of the C2-oxo group, in bacteria (23). In Escherichia coli, the presence of lysidine at the first position of the anticodon allows for AUA-decoding tRNAIle2 to recognize A and not G in the third position of the codon (26). Agmatidine has a structure very similar to lysidine in which agmatine (decarboxy-arginine) replaces the C2-oxo group of cytidine (Figure 3a) and, at the first position of the anticodon, performs the same function of differentiating between A and G at the third position of the codon in Haloarcula marismortui (23). The discrimination between A and G that results from the presence of lysidine or agmatidine is hypothesized to play an important role in the stabilization of potential positively charged tautomeric structures at neutral pH, which are able to selectively base pair with A (23).
Table 2.
Fourteen modifications have been added to the database since the last update was published in 1999
| Entry | Symbol | Common name | Phylogenetic source | RNA type | References |
|---|---|---|---|---|---|
| 96 | m1Gm | 1,2′-O-dimethylguanosine | Archaea | tRNA | (11,12) |
| 97 | m1Am | 1,2′-O-dimethyladenosine | Eukarya | tRNA | (13,14) |
| 98 | tm5U | 5-taurinomethyluridine | Eukarya | tRNA | (15,16) |
| 99 | tm5s2U | 5-taurinomethyl-2-thiouridine | Eukarya | tRNA | (15,16) |
| 100 | imG-14 | 4-demethylwyosine | Archaea | tRNA | (17,18) |
| 101 | imG2 | isowyosine | Archaea | tRNA | (17) |
| 102 | ac6A | N6-acetyladenosine | Archaea | tRNA | (19,20) |
| 103 | inm5U | 5-(isopentenylaminomethyl)uridine | Bacteria | tRNA | (21) |
| 104 | inm5s2U | 5-(isopentenylaminomethyl)-2-thiouridine | Bacteria | tRNA | (21) |
| 105 | inm5Um | 5-(isopentenylaminomethyl)-2′-O-methyluridine | Bacteria | tRNA | (21) |
| 106 | m2,7Gm | N2,7,2′-O-trimethylguanosine | Archaea | tRNA | (21) |
| 107 | m42Cm | N4,N4,2′-O-trimethylcytidine | Archaea | tRNA | (22) |
| 108 | C+ | agmatidine | Archaea | tRNA | (23) |
| 109 | m8A | 8-methyladenosine | Bacteria | rRNA | (24,25) |
Figure 3.
The two most recent additions to The RNA Modification Database: (a) agmatidine and (b) 8-methyladenosine, have been discovered and structurally characterized within the past year.
Similar to 2-methyladenosine (m2A) (27), 8-methyladenosine (m8A, Figure 3b) is present in the large subunit of bacterial ribosomes, specifically in the peptidyl transferase center at position A2503 of 23S rRNA as confirmed by comparison to the chemically synthesized mononucleoside (24). Whereas the m2A chemistry shows only a small amount of antibiotic resistance (28), the m8A moiety confers extensive resistance to five major classes of peptidyl transferase-targeting antibiotics (29). Interestingly, the m8A mononucleoside was also synthesized in 1993 and shown to be a potent inhibitor of vaccinia virus (25). This modification is a clear example of why it is important to understand the purpose of RNA modifications in living systems and how they can be utilized for therapeutic purposes. As an interesting side note, the presence of dimethylated A2503 (m2m8A) was also reported (24); however, this modification has yet to be fully characterized or structurally confirmed.
THE RNA INSTITUTE TAKES OVER AS CURATOR
As of January 2009, upkeep and supervision of the RNAMDB has been turned over to The RNA Institute. Already, this transfer has resulted in a major layout makeover in May 2009 to match the theme of the newly established institute. As a part of its mission, The RNA Institute will be expected to become a proposed hub of cutting edge RNA research and a leading provider of structural and functional information in the field of RNA modifications. For this reason, The RNA Institute will provide the logical base for new enhancements aimed at expanding the focus of the current infrastructure to cover other important aspects of modern RNA research. These enhancements will be embodied by the proposed creation of a portal that will constitute a primary resource for researchers across the entire spectrum of RNA-related disciplines. The portal will not only include the current RNAMDB and associated tools, but from this new infrastructure, users will be able to access other informatics resources for RNA science, including different repositories of experimental protocols, force field parameters for molecular dynamics simulations (30), and sister databases dedicated to non-natural RNA modifications. The infrastructure for all of the above-mentioned improvements will be implemented on a rolling basis as they are completed during 2011. A full-featured RNA portal is a goal to be reached in 2012.
The experimental protocols made available by the RNAMDB portal will comprise established procedures that are common practices in a typical RNA lab, but will also include new cutting-edge methods developed by the broader RNA community. The RNA Institute will be expected to play an important role in populating this section, due to the research interests of its members and the nature of their collaborations, which involve the development of enabling technologies based on a wide range of experimental approaches, including nuclear magnetic resonance, X-ray crystallography, single-molecule spectroscopy and, with the recent opening of The RNA Mass Spectrometry Center, high-resolution mass spectrometry. However, any investigator engaged in RNA research will be encouraged to share his/her favorite techniques with the broader RNA community through the portal. Any new protocol will require proof, either through publication in a peer-reviewed journal or empirical representative data that demonstrate its usefulness, effectiveness and reproducibility. Links to other repositories of information on specific methods, such as peer-reviewed scientific journals, will allow users to conveniently access the most up-to-date and well-characterized methods in the literature. While it is important to note that this enhancement will take a great deal of time to populate in the next 2 years with useful protocols from the various disciplines within the RNA community, the initial steps for incorporation into the RNAMDB are currently in progress.
The proposed portal will be hosting a number of tools for predicting RNA secondary structure folding, the energy of those structures, and their predicted biophysical properties, such as melting temperature. For instance, the popular RNA folding programs Mfold (31) and UNAfold (32) developed by Dr Michael Zuker will soon be accessible through the current RNAMDB infrastructure and will be subsequently incorporated in the portal. Full integration will be achieved by making the information stored in the RNAMDB directly accessible by the folding algorithms in such a way as to enable users to calculate the possible effects of RNA modifications on energetics and structure stability. Since a drastic restructuring of the source code will be required, full integration of Dr Zuker’s folding programs will likely be implemented during the 2012–13 academic year; however, the programs are currently included, in their previous state, as a part of the RNAMDB. In a similar fashion, we plan on modifying existing software to enable the productive utilization of the information in the RNAMDB. For example, full integration with the programs in the existing Masspec Toolbox suite will greatly benefit the MS identification and characterization of natural RNAs extracted from living organisms. The new tools developed for the portal will be designed to take advantage of the information stored in the site and, thus, will be capable of accessing the different types of data and seamlessly communicating with one another across the board.
The proposed portal will include a constantly updated compilation of hyperlinks to external sources that may provide additional information for the identification and the structure–function investigation of RNA. In addition to repositories of experimental protocols, these links will direct users to pertinent peer-reviewed journals, the RNA Society, the Transfer RNA Database (33), the Collaboratory for MS3D (34) and many others. Links will also smoothly point users to the external primary sources of information contained in the RNAMDB website. For instance, users will no longer be required to copy and paste the Chemical Abstracts registry numbers from the RNAMDB to the CAS Registry website. Instead, they will simply need to click on the hyperlinked CAS Registry number. Similarly, all references to both internal and external sources will be hyperlinked to provide quick access to all primary information. Additionally, it has become apparent that the information contained within the RNAMDB can be supplemented by cross-referencing with other relevant information residing in other databases such as Modomics (1) and tRNAdb (33).
Other planned enhancements to the RNAMDB functionalities include incorporation and cross-referencing with new sister databases containing the growing amount of information on both natural and non-natural RNA modifications. A sister database of force field parameters for molecular dynamics simulations will also be included as a supplement to the newly incorporated table of modified nucleoside base pairing free energy (ΔG) minima (35). Although excluded from the current RNAMDB, non-natural modifications are gaining increasing interest in both academic and corporate settings. These types of modifications include those used as research tools because of their ability to fluoresce (36), to restrict certain geometries, including backbone alterations (37), to enable unusual base pairing schemes (38) and to act as tools for probing structure (39,40). Furthermore, other non-natural nucleosides that have been discovered as the products of interactions with pharmaceuticals and as a result of environmental stimuli will be also included.
Finally, it is becoming apparent that a more modern user interface will be needed to accommodate these new enhancements. With recent technological improvements, such as deep transcriptome analysis (41) and high-throughput mass spectrometry (42,43), which have led to advancements in our current understanding of RNA in biological systems, it has become necessary to create a more automated method for researchers to submit their findings to the RNAMDB for consideration. The new user interface is being developed for implementation during 2011 and will seek to create a more user-friendly experience while adding improved functionality. The new interface will allow the curators to dedicate more of their time and resources towards validating entries and developing new features. In this direction, while past iterations allowed for unpublished modifications to be included (Table 2), the new implementation and curating structure will require publication as a criterion for entry into the RNAMDB. This will create a benchmark for the inclusion of modifications that have been diligently characterized and will reduce the likelihood of inaccurate entries.
AVAILABILITY
The RNA Modification Database is freely available to researchers via the web address: http://rna-mdb.cas.albany.edu/RNAmods/. Additionally, the programs contained in the Masspec Toolbox are also free for public use through the link on the database homepage and via the web address: http://rna-mdb.cas.albany.edu/RNAmods/rnamass.htm. Users of information contained within this database are requested to cite this article as their source of information.
FUNDING
Funding for open access charge: National Science Foundation and The RNA Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful for the contributions of Phil Durant for the 3D graphic of yeast tRNAPhe on the homepage and Patrick Limbach for his extensive work on the initial assembly and publication of the database. Additionally, we would like to thank Richard Walker, Steven Pomerantz and Byron Bossenbroek who contributed to the initial compilation of the database. The electronic version of the database was initially designed by Di He, Joe Zhang and Nancy Lombardo (University of Utah). Maintenance of the RNAMDB is supported by The RNA Institute at the University at Albany.
REFERENCES
- 1.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curran JF. Modification and editing of RNA. In: Grosjean H, Benne R, editors. Modified Nucleosides in Translation. Washington, DC: ASM Press; 1998. pp. 493–516. [Google Scholar]
- 3.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCloskey JA, Rozenski J. The Small Subunit rRNA Modification Database. Nucleic Acids Res. 2005;33:D135–D138. doi: 10.1093/nar/gki015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama S, Nishimura S. tRNA: structure, biosynthesis, and function. In: Söll D, RajBhandary U, editors. Modified Nucleosides and Codon Recognition. Washington, DC: ASM Press; 1995. pp. 207–223. [Google Scholar]
- 7.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Reports. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustilo EM, Vendeix FA, Agris PF. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCloskey JA, Liu XH, Crain PF, Bruenger E, Guymon R, Hashizume T, Stetter KO. Posttranscriptional modification of transfer RNA in the submarine hyperthermophile Pyrolobus fumarii. Nucleic Acids Symp. Series. doi: 10.1093/nass/44.1.267. Ser. No. 44, 267–268. [DOI] [PubMed] [Google Scholar]
- 12.Pettit G, Yamauchi K, Einck J. Selective Methylation of Nucleosides1. Synth. Comm. 1980;10:25–35. [Google Scholar]
- 13.Raviprakash KS, Cherayil JD. 2′-O-methyl-1-methyl adenosine: a new modified nucleoside in ragi (Eleusine coracana) tRNA. Biochem. Biophys. Res. Comm. 1984;121:243–248. doi: 10.1016/0006-291x(84)90713-7. [DOI] [PubMed] [Google Scholar]
- 14.Robins M, MacCoss M, Lee A. Nucleic acid related compounds. 20. Sugar, base doubly modified nucleosides at the 5'-terminal “cap” of mRNAs and in nuclear RNA. Biochem. Biophys. Res. Comm. 1976;70:356–363. doi: 10.1016/0006-291x(76)91053-6. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Wada T, Saigo K, Watanabe K. Novel taurine-containing uridine derivatives and mitochondrial human diseases. Nucleic Acids Res. 2001;(Suppl. 2001):257–258. doi: 10.1093/nass/1.1.257. [DOI] [PubMed] [Google Scholar]
- 16.Wada T, Shimazaki T, Nakagawa S, Otuki T, Kurata S, Suzuki T, Watanabe K, Saigo K. Chemical synthesis of novel taurine-containing uridine derivatives. Nucleic Acids Res. 2002;(Suppl. 2002):11–12. doi: 10.1093/nass/2.1.11. [DOI] [PubMed] [Google Scholar]
- 17.Zhou S, Sitaramaiah D, Noon KR, Guymon R, Hashizume T, McCloskey JA. Structures of two new “minimalist” modified nucleosides from archaeal tRNA. Bioorg. Chem. 2004;32:82–91. doi: 10.1016/j.bioorg.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Kasai H, Goto M, Ikeda K, Zama M, Mizuno Y, Takemura S, Matsuura S, Sugimoto T, Goto T. Structure of wye (Yt base) and wyosine (Yt) from Torulopsis utilis phenylalanine transfer ribonucleic acid. Biochemistry. 1976;15:898–904. doi: 10.1021/bi00649a027. [DOI] [PubMed] [Google Scholar]
- 19.Sauerwald A, Sitaramaiah D, McCloskey JA, Söll D, Crain PF. N6-Acetyladenosine: a new modified nucleoside from Methanopyrus kandleri tRNA. FEBS Lett. 2005;579:2807–2810. doi: 10.1016/j.febslet.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Aritomo K, Wada T, Sekine M. Alkylation of 6-N-acylated adenosine derivatives by the use of phase transfer catalysis. J. Chem. Soc., Perkin Trans. 1995;1:1837. [Google Scholar]
- 21.McCloskey JA, Van Wagoner RM, Hashizume T, Nomura M, Sako Y. 2003. 20th International tRNA Workshop. Banz, Germany, 41. [Google Scholar]
- 22.Nyilas A, Chattopadhyaya J. Synthesis of O2′-methyluridine, O2′-methylcytidine, N4,O2′-dimethylcytidine and N4,N4,O2′-trimethylcytidine from a common intermediate. Acta Chem. Scand. B: Org. Chem. Biochem. 1986;40:826–830. doi: 10.3891/acta.chem.scand.40b-0826. [DOI] [PubMed] [Google Scholar]
- 23.Mandal D, Köhrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Söll D, RajBhandary UL. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. PNAS. 2010;107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Aerschot AA, Mamos P, Weyns NJ, Ikeda S, De Clercq E, Herdewijn PA. Antiviral activity of C-alkylated purine nucleosides obtained by cross-coupling with tetraalkyltin reagents. J. Med. Chem. 1993;36:2938–2942. doi: 10.1021/jm00072a013. [DOI] [PubMed] [Google Scholar]
- 26.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 27.Kowalak JA, Bruenger E, McCloskey JA. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J. Biol. Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- 28.Toh S, Xiong L, Bae T, Mankin AS. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aduri R, Psciuk BT, Saro P, Taniga H, Schlegel HB, SantaLucia J. AMBER force field parameters for the naturally occurring modified nucleosides in RNA. J. Chem. Theory Comput. 2007;3:1464–1475. doi: 10.1021/ct600329w. [DOI] [PubMed] [Google Scholar]
- 31.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 33.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu ET, Hawkins A, Kuntz ID, Rahn LA, Rothfuss A, Sale K, Young MM, Yang CL, Pancerella CM, Fabris D. The collaboratory for MS3D: a new cyberinfrastructure for the structural elucidation of biological macromolecules and their assemblies using mass spectrometry-based approaches. J. Proteome Res. 2008;7:4848–4857. doi: 10.1021/pr800443f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vendeix FA, Munoz AM, Agris PF. Free energy calculation of modified base-pair formation in explicit solvent: a predictive model. RNA. 2009;15:2278–2287. doi: 10.1261/rna.1734309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nutiu R, Li Y. Aptamers with fluorescence-signaling properties. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Zhou C, Chattopadhyaya J. The synthesis of therapeutic locked nucleos(t)ides. Curr. Opin. Drug Discovery Dev. 2009;12:876–898. [PubMed] [Google Scholar]
- 38.Bergstrom DE. Unnatural nucleosides with unusual base pairing properties. Curr. Prot. Nucleic Acid Chem. 2009 doi: 10.1002/0471142700.nc0104s37. Chapter 1, Unit 1.4. [DOI] [PubMed] [Google Scholar]
- 39.Fabris D, Yu ET. Elucidating the higher-order structure of biopolymers by structural probing and mass spectrometry: MS3D. J Mass Spectrom. 2010;45:841–860. doi: 10.1002/jms.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu ET, Zhang Q, Fabris D. Untying the FIV frameshifting pseudoknot structure by MS3D. J. Mol. Biol. 2005;345:69–80. doi: 10.1016/j.jmb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 41.Iida K, Jin H, Zhu J. Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genomics. 2009;10:155. doi: 10.1186/1471-2164-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007;425:1–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 43.Kellersberger KA, Yu ET, Merenbloom SI, Fabris D. Atmospheric pressure MALDI-FTMS of normal and chemically modified RNA. J. Am. Soc. Mass Spec. 2005;16:199–207. doi: 10.1016/j.jasms.2004.10.008. [DOI] [PubMed] [Google Scholar]