Abstract
Genome-wide association studies often incorporate information from public biological databases in order to provide a biological reference for interpreting the results. The dbSNP database is an extensive source of information on single nucleotide polymorphisms (SNPs) for many different organisms, including humans. We have developed free software that will download and install a local MySQL implementation of the dbSNP relational database for a specified organism. We have also designed a system for classifying dbSNP tables in terms of common tasks we wish to accomplish using the database. For each task we have designed a small set of custom tables that facilitate task-related queries and provide entity-relationship diagrams for each task composed from the relevant dbSNP tables. In order to expose these concepts and methods to a wider audience we have developed web tools for querying the database and browsing documentation on the tables and columns to clarify the relevant relational structure. All web tools and software are freely available to the public at http://cgsmd.isi.edu/dbsnpq. Resources such as these for programmatically querying biological databases are essential for viably integrating biological information into genetic association experiments on a genome-wide scale.
INTRODUCTION
Integrating information from biological databases into high-throughput experiments, such as genome-wide association studies (GWAS), requires a database management system (DBMS) that is capable of handling very high volume and that is equipped with resources for dealing with the complex relational structure commonly seen in these databases. For example one strategy that can be used after a GWAS when selecting single nucleotide polymorphisms (SNPs) for further research is to preferentially target SNPs with evidence of biological relevance (1,2). If a SNP resides in a gene from a pathway theorized to be relevant to the phenotype or if there is evidence that the SNP has a non-neutral effect on gene expression, this biological information may increase the priority for further study of the SNP, such as additional genotyping in a replication sample. The implementation of such a strategy requires (i) direct programmatic access to biological databases in order to efficiently and viably implement the strategy on a genome-wide scale and (ii) a systematic method of isolating the relevant information within the complex network of objects and relationships within these databases. Ideally, we should also require (iii) methods for identifying the specific sequence of experiments that produced this information and for tracing these experiments back to their core biologics and original organisms in order to establish the credibility of the database and viably assess the reliability of the information being retrieved. This work is a description of our efforts to develop tools for utilizing the dbSNP relational database that meet these criteria.
The dbSNP database (3,4) is a repository that accepts submissions of data for SNPs and other structural variation such as short deletions and insertions for a multitude of organisms. It provides mapping data onto a number of conventional genomes, such as the human reference genome GRCh37 (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human), as well as mapping and functional information for gene transcripts. The database has a complex relational structure and through the tracking of submitted data users can retrieve detailed information on the experiments that led to the discovery of the variants, as well as genotype data in certain populations. dbSNP has recently been substantially expanded by data from the 1000 Genomes project (5).
Any algorithm incorporating information from a biological database on a genome-wide scale will benefit substantially from the implementation of the database using a high performance DBMS such as MySQL (http:/www.mysql.com), PostgreSQL (http://www.postgresql.org/), MSSQL (http://www.microsoft.com) or Oracle (http://www.oracle.com). Direct programmatic access to a high quality DBMS provides critical support for efficiently executed complex queries to very large databases, where a properly constructed index can mean the difference between a query taking days and minutes. The use of a conventional query language, such as SQL (6), should be used to maintain consistency across independent databases and facilitate cross-database integration, which is common practice when integrating biological information into a GWAS.
Many of the common biological databases found on the internet, such as dbSNP (3,4,7) and the University of California at Santa Cruz (UCSC) genome browser database (8), allow users to download the data in a format conducive to creating a local version of the relational database. dbSNP provides the data in a MSSQL (Microsoft) format and UCSC provides schemas compatible with MySQL. UCSC offers a small number of tables related to dbSNP containing only summary information, excluding some key information such as details on samples used and experimental methodology. It allows users to directly query their MySQL database over the internet, while dbSNP does not.
Querying these databases over the internet is not a viable solution due to the quantity of data involved. The transfer of such large amounts of data over the internet would substantially increase query execution time and place a very heavy burden on public servers. We have developed tools for downloading and implementing a local copy of the dbSNP relational database using the freely available MySQL DBMS. That is the local version exists on the user's machine so that queries to the database are done locally rather than over the internet. While dbSNP provides numerous online tools for querying and visualizing the database, as well as a download facility for retrieving the database in Microsoft MSSQL format, we have supplemented these tools with our own software for downloading and constructing a local MySQL relational database implementation of dbSNP for a specified organism. Because converting from MSSQL to MySQL is not straightforward, and the file system used in the dbSNP download facility is quite complex, our software greatly simplifies the task of implementing the relational database on a local machine. Our software was developed and tested on the Linux platform. Because it was developed using the Perl programming language, it should be readily portable to other platforms such as Microsoft Windows. This suite of tools and the conventional and freely available MySQL DBMS will allow programmers to implement complex algorithms using a fast, structured interface to the database, making the execution of these algorithms on a genome-wide scale much more viable.
While many public biological databases provide programmers with a means of creating their own local copy, this method of usage is often overshadowed by other means of data access, such as web-based query and visualization tools and proprietary application programmatic interfaces (APIs) such as those offered by Ensembl (9). The preference for these alternatives can result in less documentation and resources provided for recreating the internal relational database, making it more difficult for programmers to understand the nature of the objects and their relationships in the database. The objects of particular importance are the biologics, the experiments and the results of these experiments. The relational model is well suited to represent the basic flow of information resulting from these experiments thereby making it easier to assess the quality and reliability of the information being retrieved. More sophisticated models may be necessary to capture the entire spectrum of experimental metadata (10), although this is not necessary for some applications such as integrating biological information into a GWAS.
We have embraced the relational model as a means of elucidating the complex relational structure of the dbSNP database. We have divided the database into groups of tables corresponding to specific tasks we wish to accomplish and provide entity-relationship (ER) diagrams in the supplementary data for each task. The task model clarifies key parts of the database and suggests specific queries that should be used to accomplish each task. The ER diagrams illustrate the relationships between task-related tables and aid in the construction of queries. The task model and ER diagrams also provide the user with a clearer view of the sequence of experiments that led to the data and a means of querying specific information on the nature of those experiments.
To make these resources available to a wider audience we have developed a simple web-based query tool (http://cgsmd.isi.edu/dbsnpq) that is integrated with a database documentation tool (http://cgsmd.isi.edu/dbdoc) allowing users with a wide range of backgrounds to perform a variety of tasks with the dbSNP relational database. All tools and software are freely available to the public.
METHODS AND TOOLS
A task-based representation of dbSNP
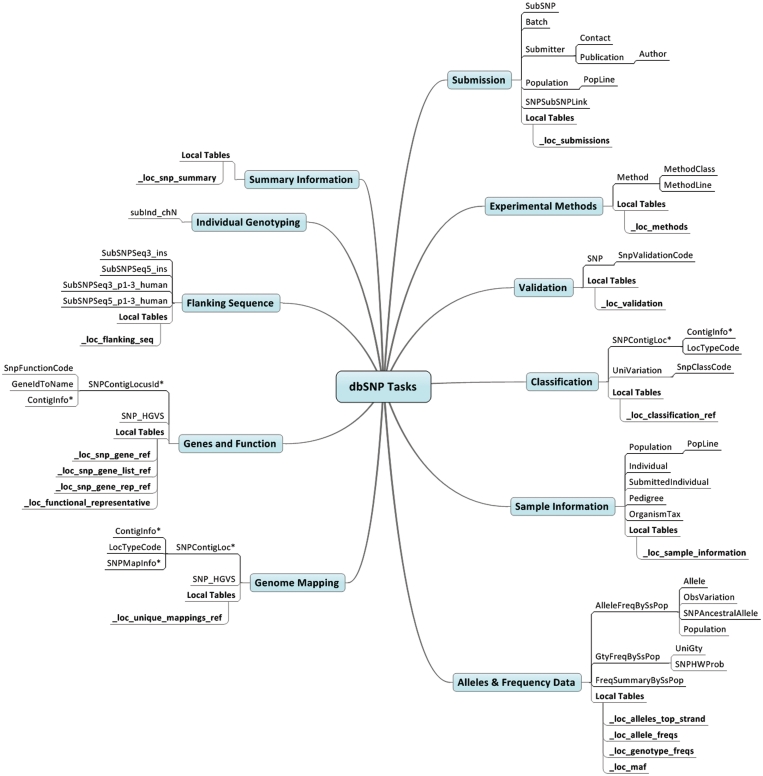
Table 1 describes the tasks we wish to accomplish with the dbSNP relational database. We use the task concept because in practice the goal is not necessarily to retrieve the full spectrum of information available, but is instead to retrieve only the portion relevant to a specific application, such as cross referencing the results of a GWAS. Figure 1 shows the relationship between our tasks and the tables in the dbSNP database (see Supplementary Table S1 for descriptions of the dbSNP tables) and illustrates some of the relational structure among the tables. The tables were downloaded from the dbSNP FTP server (ftp://ftp.ncbi.nlm.nih.gov) and correspond to build 131 of the human component of the database. In the Supplementary data we have provided a more detailed description of each task, including ER diagrams showing the schemas and relationships for the relevant tables and sample queries with output. We attempted to define the tasks so that the number of related tables is manageable and leads to interpretable ER diagrams. While we developed these methods and tools for human applications, because the database structure is consistent across different organisms, our system should translate to the other organisms maintained by dbSNP with minor modifications.
Table 1.
Descriptions of the tasks used in our classification scheme for the tables in the dbSNP database
| Task | Description |
|---|---|
| Submission | Determine the source of the submission such as specific laboratories or researchers, the populations used, any associated publications and how the submissions cluster into ‘reference SNP’ identification numbers (a group of ‘ss’ SNP IDs correspond to a unique ‘rs’ ID via the table SNPSubSNPLink). |
| Experimental methods | Determine the experimental methods used to produce the data, such as direct DNA sequencing, DNA hybridization and DHPLC (denaturing high pressure liquid chromatography). |
| Validation | Assess the reliability of the information and evaluate whether or not a reported variant is truly a genetic polymorphism or is just an experimental artifact. Methods include determining if there are multiple submissions with at least one non-computational observation and confirmation by observation of positive frequency in a genotyped sample. |
| Classification | Determine if the variants are classified as being a true SNP, insertion, deletion and so on. |
| Sample information | Retrieve information on the biological samples used, such as ethnicity and the number of samples used for a submission. |
| Alleles and frequency data | Retrieve the alleles observed for the variant, which DNA strand was used and the frequencies of the alleles and genotypes in various populations. |
| Genome mapping | Retrieve information on how the variants map to various reference genomes, such as the physical mapping coordinates and the quality of the alignments. |
| Genes and function | Retrieve information on relationships between the variants and genes, such as SNP/gene transcript functional properties (missense mutations, frameshifts, UTR regions and so on). |
| Flanking sequence | Retrieve the flanking DNA sequences used to define the variant. This can be useful when conducting custom genotyping experiments for variants not represented by commercial SNP microarrays. |
| Individual genotyping | Retrieve submitted individual genotypes. |
| Summary information | Retrieve summary information for a reference SNP ID—an amalgamation the tasks above. |
Figure 1.
The tasks and corresponding dbSNP tables from our classification scheme. A tree structure is used to partially represent the relationships between the tables. All tables except those listed under ‘Local Tables’ are directly from dbSNP. Tables with asterisks have names in the dbSNP database that are prefixed by the dbSNP build and suffixed by the representative genome build, such as b131_ContigInfo_37_1 in build 131 of the human database.
For each task we designed a small set of tables which we refer to as ‘local tables’ (Supplementary Table S3). We derived these from the original dbSNP tables in order to facilitate the tasks by providing the relevant information in a more conducive form. These local tables are often denormalized in the sense that certain numeric foreign keys are replaced by their text values and duplicate keys are replaced by comma-separated lists. While this may lead to inefficient storage the benefit is faster access to the information, which can be crucial when using the database for genome-wide analysis. Another option is to use views instead of physical tables. A view acts like a physical table but is really a query to one or more existing tables and therefore occupies no additional space. Some of our local tables have too complex a derivation to be implemented as a view and for others the queries to a view take significantly longer than to a physical table, particularly when the physical table is implemented with special indexes that improve the performance of the query. We believe the performance increase outweighs the issue of increased storage and therefore tend to use physical local tables rather than views.
A routine application of the dbSNP database to a GWAS experiment is to retrieve basic annotation, such as from our table _loc_snp_summary, for an entire commercial SNP microarray (see the section ‘Summary Information’ in the Supplementary data for details on the table _loc_snp_summary). For dbSNP human build 131 and the Illumina 1 M SNP microarray (http://www.illumina.com), which contains one million SNPs, this results in 143 million bytes of data. This is an unwieldy amount of information to query over the internet and illustrates the need for direct access to this information on a local machine. A more substantial query would be to retrieve dbSNP allele frequency data for the Illumina 1 M array via our local table _loc_allele_freqs which results in 799 million bytes of data.
The examples in the Supplementary data provide a snapshot of the flow of information in the dbSNP database and the precise types of data being stored. These examples follow the tasks in Figure 1, starting with Submission and proceeding clockwise. These represent a typical cycle of experiments and applications, from the discovery of a variant to confirmation by genotyping in multiple samples followed by genome mapping properties and gene transcript analysis and ending with representative flanking sequence to be used for additional genotyping experiments such as disease mapping studies. Our examples convey the relational structure of the database through ER diagrams and provide a detailed view of the data through example queries with output tables.
Software for downloading and implementing dbSNP
We have developed software to automate the process of downloading data and schema files from dbSNP, converting the dbSNP MSSQL schema files to MySQL and loading the data into a local MySQL server. The software is written in the Perl programming language and is executed via a UNIX command line using the syntax dbsnp.pl [command] [options]. Supplementary Table S4 lists the commands and their descriptions (see the section titled ‘Download Script’ in the Supplemenary data). The script is freely available to the public as part of the ‘dbSNP Downloader’ package which can be obtained at http://cgsmd.isi.edu/dbsnpq/downloads.php. We also provide MySQL ‘dumps’ of the human dbSNP database either as a single file containing the entire database or as a separate file for each table. These files are easily imported into a local MySQL database. The set of tables for which we provide downloads is not comprehensive—the remaining tables can be implemented using dbsnp.pl.
A web-based query tool
We have developed a web-based tool, dbSNP-Q (http://cgsmd.isi.edu/dbsnpq), for querying the human data from our MySQL implementation of the dbSNP relational database. This tool has a straightforward interface for entering SNPs and selecting a simple predefined query or entering a custom MySQL query. This allows users with a wide range of backgrounds to accomplish a variety of tasks. In addition to SNPs, users may enter genes and genomic regions in order to retrieve all the corresponding SNPs. There is also an option to look up and add all SNPs in linkage disequilibrium (LD) with the entered SNPs using one of eleven HapMap Phase III populations. Queries may be viewed directly in the browser or downloaded in Excel or tab-delimited text format. The jQuery JavaScript framework (http://www.jquery.com) ensures the interface is browser-independent and is a true interactive web-based application rather than a continuously re-loaded web page. dbSNP-Q is a model-view-controller (MVC) application: it uses Ajax technology as implemented by the jQuery plug-in jqGrid (http://www.trirand.net) to enable users to viably work with 10 s of millions of SNPs because the core logic, such as sorting and paging through the results of the query, is handled by our server. We have also implemented a vertical view technique that is useful for tables with many columns: clicking on a row immediately displays a separate table showing the contents of only that row as a two-column table.
A web-based database documentation tool
We have developed a web-based tool, dbDoc (http://cgsmd.isi.edu/dbdoc), that provides documentation for any MySQL database. The tool provides documentation and summary information on databases, individual tables, individual columns and, when available, relationships between tables and columns. It uses a web-based hierarchical navigation system to explore the database starting with a complete list of tables which can then be navigated down to documentation at the individual column level. It also provides support for grouping tables into categories identified by ‘tags’; in the case of dbSNP we use the tasks described in Table 1 (http://cgsmd.isi.edu/dbdoc/db.php?db=dbsnp_human_131#tags). In comparison, the dbSNP online data dictionary (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_db_list_table.cgi) requires users to search for tables and columns. This excludes the display of a complete list of the tables, which we believe is very useful for understanding the content of the database. dbDoc includes a search tool equipped a word completion feature making it easier for users to find documentation.
dbDoc is integrated into our query tool dbSNP-Q. When a user selects a predefined query in dbSNP-Q, a table showing documentation on each of the columns is displayed beneath the query results. The column descriptions are linked back to dbDoc so the user may browse additional documentation.
DISCUSSION
One of the themes in the development of these tools is the ability to establish the credibility of a biological database and assess the reliability of the information being retrieved from it by providing a means of tracing the information through the sequence experiments that generated the information, ideally back to the original biologics and organisms. We feel it is important to obtain this information if the data is being used to guide other experiments, such as post-GWAS prioritization of follow up studies. We also believe the information should be provided in such a way that it can be programmatically incorporated into an application such as GWAS prioritization through a DBMS such as MySQL that uses a conventional query language and table relationship paradigm and is supported by a wide variety of programming languages such as Perl and Hypertext Preprocessor (PHP).
A case in point is the dbSNP relational database which contains information on the labs and scientists that performed the experiments, the technology and the samples used in those experiments and the methods used to map the variants to the reference genomes and determine any relationships to known gene transcripts. dbSNP provides the data and schemas for all the information in their database, as well as an online data dictionary documenting the tables (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_db_list_table.cgi) and an online Handbook (http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=handbook&part=ch5). While this is a tremendous resource to investigators using SNP data, we have developed tools that enhance this resource and support our goal of establishing a programmatic interface to biological databases and promoting credibility and reliability. The task of implementing a MySQL copy of dbSNP is difficult to accomplish manually due to the MSSQL conversion not being straightforward and the complexity of dbSNP storage system for schemas and data. Furthermore, our task-based table classification system with ER diagrams and custom local tables facilitate usage of the database. dbSNP does provide an ER diagram of their database at ftp://ftp.ncbi.nih.gov/snp/database/erd_dbSNP.pdf. At the time of writing this diagram was last updated in 2005 and some tables in the diagram are obsolete. While it does divide the tables into different subject areas, the result is still quite complex. We believe that focusing on more narrowly defined tasks leads to a more manageable set of tables and therefore a more viable resource for programmers.
The UCSC (8) and Ensembl (9) sites are two other popular sources of biological information that provide data and schemas in MySQL format as well as direct MySQL access. The tables from the UCSC human database relevant to dbSNP, for example snp130 and snp130CodingDbSnp for build 130, provide summary information similar to our _loc_snp_summary table. The Ensembl database offers additional information (http://pre.ensembl.org/info/docs/api/variation), but both sources lack data on submitters and technological methods which we believe detracts from the credibility of the database.
A limitation in our methods is determining when there are errors in the dbSNP data (7,11) as we provide no resources to specifically address this issue. Our goal is to first clarify the structure of the database in terms of identifying what it is we intend to do with the information, hence the task-based representation. The next steps in developing tools that utilize databases such as dbSNP should be to determine whether it is necessary to incorporate additional validation mechanisms and then implement these mechanisms in such a way that any quality-control procedures that affect the data are clearly traceable, in the same way we trace the experiments that produced the data.
An additional service that may be provided in future implementations of these tools is the provision of probe sequence data from commercial SNP microarrays such as those offered by Affymetrix (http://affymetrix.com) and Illumina (http://illumina.com). While we do provide the dbSNP flanking sequence data that was originally reported by submitters, it may be useful to obtain probe sequence data directly from the manufacturers and provide this data to investigators using similar relational database methods. One application would be the resolution of strand-ambiguous genotype calls.
We have developed a systematic workflow for retrieving and processing data from dbSNP and implementing a local MySQL relational database. This workflow will provide a robust solution for working with future releases of dbSNP. The primary utility for automating this workflow is our dbSNP Downloader package that is freely available to the public at http://cgsmd.isi.edu/dbsnpq/downloads.php. Future releases may involve changes to the format and structure of the files on the dbSNP download site, such as changes to dbSNP database schema files and we will modify our software to accommodate these changes. We have found that these changes may sometimes be subtle and yet have serious consequences, such as a text file that suddenly uses Windows line delimiters when the format was UNIX in the previous download from dbSNP. This highlights the importance of having a robust system for downloading and processing data. Our dbSNP Downloader package system will enhance our ability to ensure the dbSNP-Q web tool provides the most recent build of human dbSNP data.
In order to allow these resources to broaden and remain useful to the general scientific community rather than only to programmers, we will expand our dbSNP-Q and dbDoc web tools and accompanying software packages such as dbSNP Downloader to include experimental genomic data from sources other than dbSNP. This expansion would be subject to the limitations dictated by our goal of establishing credible relational databases of information that can be traced back through specific experiments and processes to the original subjects and biologics. For example it is not clear if it is practical to develop new utilities for implementing the complete UCSC and Ensembl databases. One reason is that these sites in particular already provide resources for implementing local versions of their databases using software such as MySQL. Another reason is that the information provided by these sites is often derived from other sources such as dbSNP. Our goal, as in the case of dbSNP, is to develop databases for specific experimental sources using the traceability criteria to establish credibility. Nevertheless, integrating these extensive genomic databases into a common resource that includes our dbSNP tools could improve the ability of researchers to integrate a vast array of genomic experimental data using conventional programming methods and therefore this undertaking will be explored. Another resource is the HapMap database, which is already integrated indirectly into the dbSNP-Q web tool for the purpose for querying data for all SNPs in LD with specified SNPs. We are currently designing a HapMap relational database similar to our dbSNP implementation. One difficulty is that the HapMap site (http://hapmap.ncbi.nlm.nih.gov) does not provide any form of relational database schema to model such aspects as population data and the different technologies used to produce the genotype data. Therefore these relational database models must be developed independently.
A key theme in our work is programmatic access to the database, by which we mean a DBMS that is supported by conventional programming languages so that complex algorithms can be implementing using a fast access paradigm to the database. For example we prefer the widely used Perl programming language (http://www.perl.org) along with the module Perl::DBI (http://dbi.perl.org) for MySQL support. Web software development can take advantage of the Linux-Apache-MySQL-PHP (LAMP) and Ajax programming paradigms, where MySQL-friendly PHP scripts can be used to develop web applications, such as our own SNP prioritization tool (https://spot.cgsmd.isi.edu) (2) and the dbSNP-Q query tool (http://cgsmd.isi.edu/dbsnpq). MySQL is a very widely used DBMS with freely available client and server software and numerous professional grade development tools (http://dev.mysql.com). The ability to implement local MySQL copies of large biological databases will be conducive to the development of public tools for integrating this information into other biological experiments such as GWAS.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute on Drug Abuse (K01 DA024722 to S.F.S.); American Cancer Society (IRG 5801050 to S.F.S.); National Institute on Mental Health (U24 MH068457 to J.A.T.). Funding for open access charge: National Institutes of Health grants [DA024722 (50%) and MH068457 (50%)].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to the following individuals for testing our software: Andrew Schrage, Richard McEachin and Sharon Ryan. The authors are also very grateful for the assistance received from the dbSNP administrators. The authors thank the reviewers for their comments and suggestions, which helped to improve the quality of the article. Finally, the authors thank the editor, Dr Michael Galperin, for the suggestion of making this work accessible to a wider scientific audience, which led to the development of the dbSNP-Q web tool.
REFERENCES
- 1.Saccone SF, Saccone NL, Swan GE, Madden PA, Goate AM, Rice JP, Bierut LJ. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics. 2008;24:1805–1811. doi: 10.1093/bioinformatics/btn315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saccone SF, Bolze R, Thomas P, Quan J, Mehta G, Deelman E, Tischfield JA, Rice JP. SPOT: a web-based tool for using biological databases to prioritize SNPs after a genome-wide association study. Nucleic Acids Res. 2010;38(Suppl.):W201–209. doi: 10.1093/nar/gkq513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38(Database issue):D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuehn BM. 1000 Genomes Project promises closer look at variation in human genome. JAMA. 2008;300:2715. doi: 10.1001/jama.2008.823. [DOI] [PubMed] [Google Scholar]
- 6.Jamison DC. Structured Query Language (SQL) fundamentals. Curr. Protoc. Bioinformatics. 2003 doi: 10.1002/0471250953.bi0902s00. Chapter 9, Unit 9.2. [DOI] [PubMed] [Google Scholar]
- 7.Day IN. dbSNP in the detail and copy number complexities. Hum. Mutat. 2009;31:2–4. doi: 10.1002/humu.21149. [DOI] [PubMed] [Google Scholar]
- 8.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC genome browser database: update 2010. Nucleic Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones AR, Lister AL. Managing experimental data using FuGE. Methods Mol. Biol. 2009;604:333–343. doi: 10.1007/978-1-60761-444-9_23. [DOI] [PubMed] [Google Scholar]
- 11.Musumeci L, Arthur JW, Cheung FS, Hoque A, Lippman S, Reichardt JK. Single Nucleotide Differences (SNDs) in the dbSNP Database May Lead to Errors in Genotyping and Haplotyping Studies. Hum. Mutat. 2010;31:67–73. doi: 10.1002/humu.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]