Abstract
MicroRNAs (miRNAs) represent an important class of small non-coding RNAs (sRNAs) that regulate gene expression by targeting messenger RNAs. However, assigning miRNAs to their regulatory target genes remains technically challenging. Recently, high-throughput CLIP-Seq and degradome sequencing (Degradome-Seq) methods have been applied to identify the sites of Argonaute interaction and miRNA cleavage sites, respectively. In this study, we introduce a novel database, starBase (sRNA target Base), which we have developed to facilitate the comprehensive exploration of miRNA–target interaction maps from CLIP-Seq and Degradome-Seq data. The current version includes high-throughput sequencing data generated from 21 CLIP-Seq and 10 Degradome-Seq experiments from six organisms. By analyzing millions of mapped CLIP-Seq and Degradome-Seq reads, we identified ∼1 million Ago-binding clusters and ∼2 million cleaved target clusters in animals and plants, respectively. Analyses of these clusters, and of target sites predicted by 6 miRNA target prediction programs, resulted in our identification of approximately 400 000 and approximately 66 000 miRNA-target regulatory relationships from CLIP-Seq and Degradome-Seq data, respectively. Furthermore, two web servers were provided to discover novel miRNA target sites from CLIP-Seq and Degradome-Seq data. Our web implementation supports diverse query types and exploration of common targets, gene ontologies and pathways. The starBase is available at http://starbase.sysu.edu.cn/.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous ∼22 nt RNAs that direct the post-transcriptional repression of protein-coding genes (1,2). By base pairing to mRNAs, miRNAs mediate translational repression or mRNA degradation (1–3). Functional studies indicate that miRNAs participate in the regulation of numerous cellular processes, such as proliferation, apoptosis, differentiation and the cell cycle (1–3).
Thousands of miRNAs have been identified in animals and plants by cloning and deep sequencing, but determining the targets of these miRNAs is an ongoing challenge (1–4). To date, a large number of target prediction computer programs have been developed, such as TargetScan (5,6), PicTar (7), miRanda (8), PITA (9) and RNA22 (10) for animal miRNA targets, and miRU (11) and TargetFinder (12) for plant miRNA targets. In addition, several resources have been established to systematically collect and describe both experimentally validated miRNA targets [TarBase (13), miRecords (14)] and predicted miRNA targets [miRGator (15), MiRNAMap (16)]. However, because miRNA regulation of an animal mRNA requires base pairing with only few nucleotides of the 3′-UTR region of the target mRNA, different target prediction programs produce different results and have high false positive rates (4,17–19). Although plant miRNA targets have been predicted on the basis of their extensive and often conserved complementarity to miRNAs (1,2), we must spend substantial time and effort attempting to validate predicted miRNA targets that turn out to be false.
In the past several years, significant efforts have been made in determining biologically relevant miRNA–target interactions using high-throughput experimental approaches. Several recent studies have reported the use of cross-linking and Argonaute (Ago) immunoprecipitation coupled with high-throughput sequencing [CLIP-Seq, also referred to as high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP)] (20–22) and high-throughput degradome sequencing [Degradome-Seq, also referred to as ‘parallel analysis of RNA ends’ (PARE)] (23–27) to isolate targets in animals and plants. The application of CLIP-Seq and Degradome-Seq methods has significantly reduced the rate of false positive predictions of miRNA binding sites and has also reduced the size of the search space for miRNA target sites (20–27). The increasing amount of CLIP-Seq and Degradome-Seq data generates a strong demand among researchers for an integrated database that could facilitate the annotation and analysis of these data.
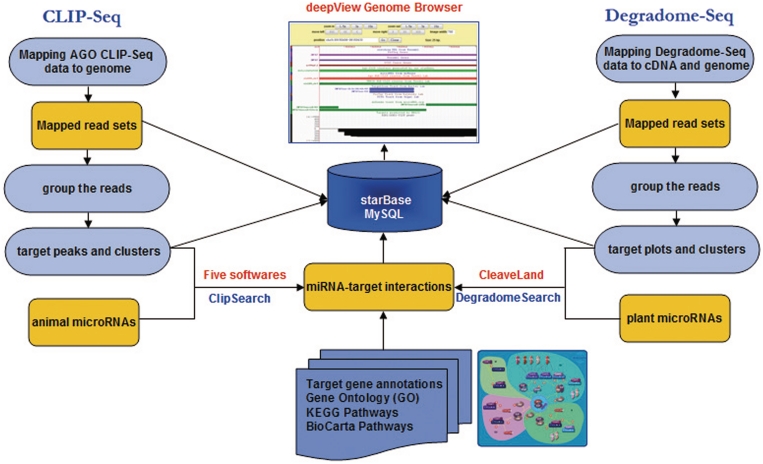
To meet this need, we have developed and are introducing via the current study, the starBase database. The starBase facilitates the integrative, interactive and versatile display of, as well as the comprehensive annotation and discovery of, miRNA–target interaction maps from CLIP-Seq and Degradome-Seq data from six organisms: human, mouse, Caenhorhabditis elegans, Arabidopsis thaliana, Oryza sativa and Vitis vinifera (Figure 1). Information on tens of thousands of miRNA–target regulatory relationships, as well as millions of Ago-binding sites and cleavage sites (Table 1) is contained within starBase. In addition, two novel web servers were developed to identify miRNA binding sites or cleavage sites from CLIP-Seq and Degradome-Seq data. As a means of comprehensively integrating Ago CLIP-Seq and Degradome-Seq data, this database is expected to provide considerable resources to help researchers investigating new miRNA–target interactions and developing next generation miRNA target prediction algorithms.
Figure 1.
System overview of starBase core framework. All results generated by starBase are deposited in MySQL relational databases and displayed in the visual browser and web page.
Table 1.
Data statistics in starBase
| Species | Library | Mapped read | Peak cluster | Relationship | Known target |
|---|---|---|---|---|---|
| Human | 13 | 1 137 137 | 390 990 | 156 751 | 536 |
| Mouse | 5 | 2 831 042 | 725 145 | 232 196 | 132 |
| Caenhorhabditis elegans | 3 | 178 639 | 24 699 | 12 904 | 53 |
| Arabidopsis | 6 | 5 575 026 | 669 589 | 25 688 | / |
| Oryza sativa | 3 | 6 448 130 | 1 009 870 | 37 254 | / |
| Vitis vinifera | 1 | 2 404 808 | 431 180 | 3179 | / |
Statistics indicating the numbers of library (CLIP-Seq and Degradome-Seq), mapped reads, peak clusters (CLIP-Seq clusters or Degradome-Seq clusters), relationships (miRNA–target regulatory relationships) and known animal miRNA target sites for the six organisms (human, mouse, C. elegans, Arabidopsis, Oryza sativa and Vitis vinifera). ‘/’ means that known plant miRNA target sites are not present in the table because miRecords (14) provides only animal miRNA target sites. It should be noted that some miRNA–target regulatory relationships involved miRNAs from the same family and involved mRNA isoforms.
MATERIALS AND METHODS
Twenty-one Ago or TNRC6 CLIP-Seq sequence data sets and 10 Degradome-Seq sequence data sets were compiled from eight related studies (20–27) and downloaded from NCBI GEO database (28) or obtained from the Supplementary Data of the original articles (20–27). Ago and TNRC6 CLIP-Seq reads were mapped to genomes using the Bowtie program (version 0.12.0) (29) with options:-a --best --strata -v 2 -m 1. Reads with multiple equivalent hits to the genome were discarded. Degradome-Seq data were mapped to genomes and cDNA sequences using the Bowtie program (version 0.12.0) (29) with options: -a -m 10 -v 0 and -a -v 0, respectively. The overlapping reads were grouped into clusters, with each cluster including at least one sequence read and having a minimum length of 20 nt. In addition, the high reliable Ago (ALG-1) CLIP-Seq clusters in L4-stage wild-type (wt) worms, the top Ago or TNRC6 CLIP-Seq clusters in human HEK293 cells and the Ago CLIP-Seq clusters/peaks in mouse neocortex were obtained from the supplementary material of the original articles (20–22).
Animal miRNA target sites predicted by 5 prediction programs: [TargetScan (5,6), PicTar (7), miRanda (8), PITA (9) and RNA22 (10)] were downloaded from their corresponding websites. Predicted target site coordinates from TargetScan (5), PicTar (7) and PITA (9) were converted to coordinates of recently released genomes using the liftOver utility from the University of California Santa Cruz (UCSC) bioinformatics websites (30). Target sequences from RNA22 (10) and miRanda (8) were aligned to genomes to determine genome coordinates using the Bowtie program (29). Plant miRNA target sites were predicted by the CleaveLand program (version 2.0) (31). We only considered the miRNA–target interactions with an alignment score from CleaveLand not exceeding the cutoff threshold of 7.0. Experimentally validated target sites were downloaded from the miRecords website (14). Then these validated target sites were mapped to genomes to allow determination of genome coordinates using the Bowtie program (29).
The ClipSearch program was developed to search for 6–8-mers (8-mer, 7-mer-m8 and 7-mer-A1) (2,5) in CLIP-Seq data. The DegradomeSearch program was developed to search Degradome-Seq clusters for nearly perfect complements of miRNA sequences. Degradome-Seq cluster sequences, extended by an additional 15 nt in both the 5′- and 3′-directions for each of the species, were extracted and used as the DegradomeSearch input data set. DegradomeSearch web-server aligned miRNA to extended clusters using segemehl (version 0.093) (32). Interactions between miRNA and target were scored according to the previously described methods (31,33,34). For each interaction, we performed a search of the genome-wide cleavage sites pre-deposited in the MySQL database to determine if there were cleavage tags at the 10th nucleotide of the alignment.
All known miRNAs were downloaded from miRBase [release 15.0, (35)]. All refGenes were downloaded from the UCSC bioinformatics websites (30). GO ontology (36), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (37) and BioCarta pathways for refGenes were extracted from UCSC Table browser (30). Known non-coding RNA genes were downloaded from Ensembl (38) or UCSC (30) or were obtained from related literature (39–41). Human (UCSC hg19), mouse (UCSC mm9, NCBI Build 37) and C. elegans (WS190) genome sequences were downloaded from UCSC bioinformatics websites (30). Arabidopsis (TAIR9) genome sequences were downloaded from TIGR (42). Rice genome sequences were downloaded from the MSU Rice Genome Annotation Website (43). Grapevine genome sequences were downloaded from the Genoscope website (44).
DATABASE CONTENT
Genomic landscape of Ago-binding sites and miRNA-cleavage sites
To study genome-wide Ago–RNA interaction patterns, we grouped ∼4 million mapped CLIP-Seq reads into about 1 million clusters (details in ‘Materials and Methods’ section, Table 1). The sequencing depth distribution of these clusters is presented in the form of target peaks (t-peaks), which were displayed in our deepView genome browser (Figure 1 and Supplementary Figure S1). This display method allows a direct comparison of peak patterns generated from different Ago proteins, cell lines and tissues to determine miRNA target sites. In general, clusters corresponding to the bona fide binding site are found at a higher peak than those corresponding to biological noise (Figure 1 and Supplementary Figure S1). All clusters were intersected with annotated genomic elements, and >10% of clusters were found to overlap known 3′-UTR regions in each species (Supplementary Table S1). Intriguingly, we found that >10% and >7% of clusters overlapped with CDS and intron regions, respectively, in each species (Supplementary Table S1).
The same strategy was applied to group ∼14 million mapped Degradome-Seq reads into about 2 million clusters (Table 1). The majority of these clusters overlapped with the mRNA sequences. To study patterns of RNA degradation, genome-wide target plots (t-plots) (23,45) (Supplementary Figure S2) were constructed by plotting the abundance of each cleavage signature on the genome sequences. As described by German et al. (23,45), these t-plots can be used to distinguish true miRNA cleavage sites from background noise. In general, for bona fide miRNA targets, the cleavage tags corresponding to the cleavage site are found at higher abundances than those at other positions, making them fairly easy to distinguish by simple observation of the t-plots (23,45) (Supplementary Figure S2).
Annotation and identification of miRNA–target regulatory relationships
To investigate animal miRNA–target regulatory relationships, animal miRNA target sites predicted by the five prediction programs [TargetScan (5,6), PicTar (7), miRanda (8), PITA (9) and RNA22 (10)] were intersected with all CLIP-Seq peak clusters. In total, we identified approximately 400 000 regulatory relationships between 1348 miRNAs and 26 296 genes (Table 1 and Supplementary Table S2). By using CLIP-Seq data to filter candidates, the predicted results of each target prediction program were significantly reduced, suggesting that there may be a number of false positive predictions generated from different computational approaches. To provide valuable insights as to the function of each miRNA, we carried out a comprehensive gene set analysis of the miRNA target sets by combining the KEGG pathways (37), the BioCarta pathways and the Gene Ontologies (GO) categories (36).
We applied the CleaveLand program (version 2.0) (31) to plant Degradome-Seq data, and identified approximately 66 000 miRNA–target regulatory relationships that involved 25 579 genes and 856 miRNAs (Table 1 and Supplementary Table S2). Due to the integration of the large number of Degradome-Seq libraries from diverse tissues, this analysis provides an enhanced resolution for these regulatory relationships.
Predicting target sites of small RNAs from CLIP-Seq and Degradome-Seq data
The increasing amount of CLIP-Seq and Degradome-Seq data also produces a strong demand for web-based tools to predict target sites of small RNAs from these data. Two web-based tools, ClipSearch and DegradomeSearch, were developed to screen the potential miRNA binding sites and cleavage sites. ClipSearch predicts biological miRNA–target interactions by searching for 8-mer and 7-mer sites that match the seed region of the miRNA. ClipSearch searches for these sites in CLIP-Seq clusters that overlap with the 3′-UTR of the known genes. ClipSearch can discover non-conserved miRNA binding sites because it does not use cross-species sequence conservation to filter candidates.
DegradomeSearch predicts functional miRNA–target interactions by searching for sites with a near-perfect match to the whole miRNA sequence. DegradomeSearch searches for these sites in Degradome-Seq clusters that overlap with mRNA (details in ‘Materials and Methods’ section). Interactions are scored according to a scoring scheme that successfully identified miRNA target sites in plants (31,33,34). In its default setting, DegradomeSearch finds miRNA–target interactions with a penalty score not exceeding 7.0 and having at least one cleavage tag. False positives or predicted results can be reduced by choosing a lower penalty score or by limiting the minimum number of cleavage tags.
WEB INTERFACE
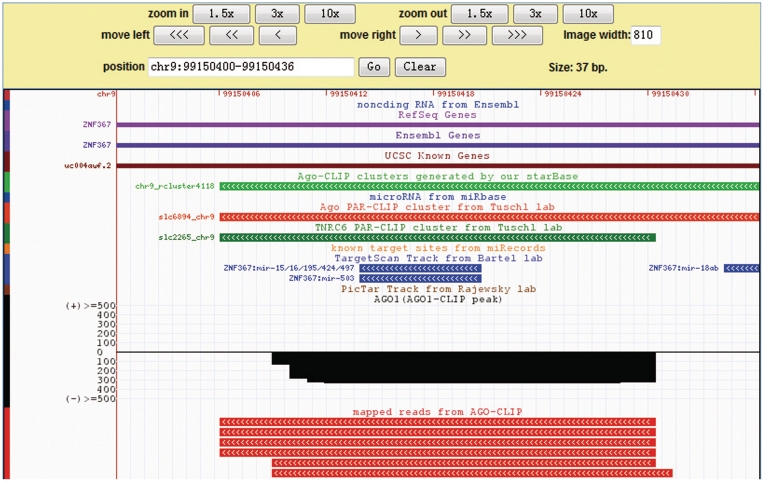
The starBase database provides various query interfaces and graphical visualization pages to facilitate analysis of the CLIP-Seq and Degradome-Seq data sets and exploration of miRNA–target interactions. Our improved deepView Genome Browser (46) provides an integrated view of mapped reads, predicted and known miRNA targets, ncRNAs, protein-coding genes, target clusters, target-peaks and target-plots (Figure 2 and Supplementary Figures S1–S2). Bench biologists can use the genome browser to simultaneously compare the maps of t-peaks or t-plots generated from multiple experiments and the conservation of binding sites from all target prediction programs. Clicking a track item within the browser launches a detailed page providing further information on that item or links to external resources such as NCBI, UCSC and TAIR, from which one can obtain more comprehensive information.
Figure 2.
Illustrative screen shots from the deepView browser. The deepView browser provides an integrated view of CLIP-Seq and Degradome-Seq data, known and predicted miRNA target sites, protein-coding genes, ncRNA genes, miRNAs, strand-specific peak clusters, genome-wide target-peaks and target-plots.
Web-based exploration of miRNA–target regulation relationships
We provide two web interfaces, CLIP-Seq and Degradome-Seq, with which to display the miRNA–target interaction relationships (Supplementary Figure S3–S4). Users can browse the relationships by entering a gene name or by selecting a microRNA name. When one starts typing a gene name in the search box, suggested gene names are displayed in the list box. The user can then either choose a gene from the list box or finish typing the full gene name. The user can also search for intersections among targets by choosing interested target prediction programs. The results of the search are listed as the miRNA–target table. For the CLIP-Seq section, the number of predicted binding sites given by each prediction program and the number of CLIP-Seq reads are indicated in a table. The users can click on the number within the table to launch a detailed page providing further information on that miRNA–target interaction. The user also can click on the title of the table to sort miRNA–target interactions according to various features, such as the number of binding sites, miRNA names or gene names. The detailed information for a miRNA–target interaction includes a description of the target gene, the GO terms of the gene, the pathways the target gene is involved in and the number of Clip-Seq reads (Supplementary Figure S3). This information allows the user to filter the putative targets further.
The Degradome-Seq section is organized similarly to the CLIP-Seq section. The target genes, the genomic coordination, the penalty score of miRNA–mRNA interactions and the sequence number of cleavage sites are all presented in a table. Clicking on the target gene within the table launches a page showing detailed information on the miRNA–target interactions (Supplementary Figure S4).
Web-based tools for predicting target sites of novel small RNAs from CLIP-Seq and Degradome-Seq data
The starBase provides two simple and user-friendly interfaces to allow the users to predict target sites of small RNAs from CLIP-Seq and Degradome-Seq data (Supplementary Figures S5–S6). The user is required to select an intended organism, and then enter nucleotides 2–8 of a mature sequence or a mature miRNA sequence for the ClipSearch and DegradomeSearch programs, respectively. After data submission, a typical run may take several minutes to finish. To reduce false positives in the predicted targets from the ClipSearch program, the user can filter the candidate targets by selecting site types, which are classified into 8-mer, 7-mer-m8 and 7-mer-A1 (2,5). The user can also limit the penalty score to reduce the false positive predictions in the DegradomeSearch program. The sequence depth of a target site or cleavage site can be used to further reduce false positives in the predicted targets. The output of the ClipSearch program consists of three parts: site type, information about the target gene and visual sequence alignments matched to a specific CLIP-Seq cluster (Supplementary Figure S7). The output of the DegradomeSearch program also consists of three parts: the penalty score, the miRNA–mRNA interaction map and the sequence number of cleavage site in different experiments (Supplementary Figure S8). A link to the DeepView genome browser is also provided to allow the user to view various features of each target region.
DISCUSSION AND CONCLUSIONS
Our global analysis of Ago CLIP-Seq and Degradome-Seq data derived from 31 experiments in six organisms provides a comprehensive integrated map of the miRNA–target interactions. The large number of Ago-binding sites and cleavage sites identified in this study have shown there to be an extensive and complex interaction map among Ago proteins, miRNAs and target RNAs (Table 1).
Our initial analysis found that the majority of CLIP-Seq and Degradome-Seq clusters could not be clearly predicted to be miRNA targets (Supplementary Table 1), implying they may bind to novel small RNAs, or miRNAs that follow unexpected rules of binding, such as the centered pairings (center sites) recently reported by Bartel and his colleagues (47). Moreover, numerous CLIP-Seq clusters were not located within the 3′-UTR of the gene, indicating that the miRNA may bind to the coding region and the 5′-UTR, as has been reported for ribosomal protein regulation by mir-10a (48) and Nanog, Oct4 and Sox2 regulation by miR-134, miR-296 and miR-470 (49). Recent reports revealed that the Ago protein also plays a role in miRNA-derived cleavage (47) and in miRNA processing (50) in animals. Therefore, one might speculate that a substantial number of Argonaute-catalyzed cleavage sites may be hiding in these data. In plants, vast amounts of Degradome-Seq data might not be miRNA-derived cleavage sites, but rather the by-product of other degradation pathways. Nevertheless, we anticipate that future investigation of these data might provide important insights into rules governing miRNA–target interactions.
Compared to the other miRNA target-related databases, including TarBase (13), miRecords (14), miRGator (15) and MiRNAMap (16), which only collect predicted targets or experimentally supported targets, the distinctive features in our starBase database are as follows: (i) CLIP-Seq and Degradome-Seq are the newest high-throughput technology for the transcriptome-wide identification of miRNA target sites in animals and plants (20–27). Our starBase database is the first database to provide comprehensive analysis of public CLIP-Seq and Degradome-Seq data, (ii) genome-wide t-peak and t-plot maps generated by starBase allow users to easily search within these signatures for the miRNA cleavage sites or binding sites, (iii) our improved deepView browser in starBase provides an integrated view of multidimensional data to facilitate miRNA regulatory networks research (Figure 2, Supplementary Figures S1–S2), (iv) two web-based tools, ClipSearch and DegradomeSearch, can be used to predict animal and plant target sites of small RNAs from CLIP-Seq and Degradome-Seq data. We expect that access to these tools will enable more researchers to search for target sites of novel miRNAs or endo-siRNAs in the ever-increasing amounts of CLIP-Seq and Degradome-Seq data, (v) The starBase database also provides users the GO annotation and biological pathways of miRNA targets (Figure 1). These associated terms may provide valuable insights into the regulatory role and function of each miRNA.
We have provided a variety of information to facilitate exploration of miRNA–target interaction maps. Although some CLIP-Seq clusters with small read numbers may simply represent experimental or biological noises, users can further filter these CLIP-Seq clusters by viewing whether they overlapped with bona fide clusters obtained from the original articles, how many reads mapped to the CLIP-Seq cluster and how many CLIP-Seq experiments include the CLIP-Seq cluster. We expected that the data, web-based tools and the integrative, interactive and versatile display provided by the starBase database will aid future experimental and computational studies to discover new miRNA target sites and miRNA–target interaction features.
FUTURE DIRECTIONS
CLIP-Seq and Degradome-Seq technologies have provided powerful ways to study biologically relevant miRNA–target interactions at the transcriptome-wide level. As these technologies are applied to a broader set of species, cell lines, tissues and conditions, we will continuously maintain and update the database to keep up with these improvements. Moreover, we will continue to increase the amount of storage space and improve the computational efficiency of our computer servers for storing and analyzing these new data. In addition, we intend to integrate other CLIP-Seq data from other RNA binding proteins (51), such as PUM2 (22), Nova (52), FOX2 (53) and PTB (54), into starBase to improve our understanding of the eukaryotic regulatory networks.
AVAILABILITY
The starBase database is freely available at http://starbase.sysu.edu.cn/. All starBase data files can be freely downloaded and used according to the GNU Public License.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (No. 30830066, U0631001, 30900820); Ministry of Science and Technology of China, National Basic Research Program (No. 2005CB724600, 2011CB811300); the funds from the Ministry of Education of China and Guangdong Province (No. IRT0447, NSF05200303, 9451027501002591); China Postdoctoral Science Foundation (No. 20080440800, 200902348). Funding for open access charge: Ministry of Science and Technology of China, National Basic Research Program (No. 2011CB811300).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ling-ling Zheng for her valuable comments; Markus Hafner and Thomas Tuschl for providing the PAR-CLIP data; Robert Darnell for providing the HITS-CLIP clusters and peaks; and Gene Yeo for providing CLIP-Seq data.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 10.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahlgren N, Carrington JC. miRNA target prediction in plants. Methods Mol. Biol. 2010;592:51–57. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- 13.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36:D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 18.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 20.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 24.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. Rice MicroRNA effector complexes and targets. Plant Cell. 2009;21:3421–3435. doi: 10.1105/tpc.109.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J. Identification of grapevine microRNAs and their targets using high throughput sequencing and degradome analysis. Plant J. 2010;62:960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou M, Gu L, Li P, Song X, Wei L, Chen Z, Cao X. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica) Front. Biol. 2010;5:67–90. [Google Scholar]
- 28.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 34.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown JW, Echeverria M, Qu LH, Lowe TM, Bachellerie JP, Huttenhofer A, Kastenmayer JP, Green PJ, Shaw P, Marshall DF. Plant snoRNA database. Nucleic Acids Res. 2003;31:432–435. doi: 10.1093/nar/gkg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan AP, Rabinowicz PD, Quackenbush J, Buell CR, Town CD. Plant database resources at The Institute for Genomic Research. Methods Mol. Biol. 2007;406:113–136. doi: 10.1007/978-1-59745-535-0_5. [DOI] [PubMed] [Google Scholar]
- 43.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 45.German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- 46.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 50.Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]