Abstract
Most proteins from higher organisms are known to be multi-domain proteins and contain substantial numbers of intrinsically disordered (ID) regions. To analyse such protein sequences, those from human for instance, we developed a special protein-structure-prediction pipeline and accumulated the products in the Structure Atlas of Human Genome (SAHG) database at http://bird.cbrc.jp/sahg. With the pipeline, human proteins were examined by local alignment methods (BLAST, PSI-BLAST and Smith–Waterman profile–profile alignment), global–local alignment methods (FORTE) and prediction tools for ID regions (POODLE-S) and homology modeling (MODELLER). Conformational changes of protein models upon ligand-binding were predicted by simultaneous modeling using templates of apo and holo forms. When there were no suitable templates for holo forms and the apo models were accurate, we prepared holo models using prediction methods for ligand-binding (eF-seek) and conformational change (the elastic network model and the linear response theory). Models are displayed as animated images. As of July 2010, SAHG contains 42 581 protein-domain models in approximately 24 900 unique human protein sequences from the RefSeq database. Annotation of models with functional information and links to other databases such as EzCatDB, InterPro or HPRD are also provided to facilitate understanding the protein structure-function relationships.
INTRODUCTION
Nowadays, genome sequencing projects are producing complete genome sequences at an extremely high rate (1,2). With the rise of next-gen sequencers (3–5), this is the continuous trend for the future without a doubt. Consequently, the number of known protein sequences (6) grows more rapidly than the number of known protein structures experimentally determined (7). However, to make full use of genome sequences, proteins encoded in genomes should be analysed and for this purpose, protein three-dimensional (3D) structures provide much information (8,9). Computational methods for protein 3D structure prediction are anticipated to bridge the gap between the number of known protein sequences and the number of known protein structures. According to assessments of the accuracy of those methods, e.g. recent Critical Assessment of Techniques for Protein Structure Prediction (CASP) experiments (10,11), template-based protein structure prediction often produced 3D models accurate enough for functional annotations, modification of protein functions or even for structure-based drug design (12,13). In addition, in the CASP7 and 8 experiments, fully automated structure prediction methods had reached a comparable level to the best prediction performance by methods with human intervention (14).
In the CASP experiments, target protein sequences are ones whose 3D structures will be determined. It means that such protein structures are expected to be single domains or a couple of domains and suitable for the experimental structure determination. Therefore, sometimes protein sequences are truncated from their full-length forms. On the other hand, most protein sequences coded in genomes from higher organisms are known to be long and should be multi-domain proteins (15), and contain a significant portion of intrinsically disordered (ID) regions (16–19). Clearly, these proteins are unsuitable for experimental structure determination in the full-length form and distinct from the target protein sequences of CASPs. To analyse such proteins, we have developed a special protein-structure-prediction pipeline, by integrating and arranging various computational tools, either developed by us or widely used as global standards. This pipeline was applied to all proteins coded in the human genome. The resulting 3D models as well as other annotations for protein functions were accumulated in the Structural Atlas of Human Genome (SAHG) database and presented through the web interface at http://bird.cbrc.jp/sahg.
There are other databases of protein structure models, e.g. SWISS-MODEL Repository (20) or ModBase (21). Both databases contain annotated protein structure models generated by original automated modeling pipelines. They also allow the users to build models on demand. Compared with them, the SAHG database is distinct mainly in the following points: (i) The 3D models in SAHG were generated by an original pipeline, specific for multi-domain proteins with substantial ID regions; (ii) Conformational changes of proteins upon ligand-binding are predicted by simultaneous modeling using templates of the ligand-bound state (holo form) and the unbound state (apo form) and displayed as animated images; and (iii) Functional annotations for protein interactions, e.g. ligand-binding and protein–protein interactions, are available. All these features are suitable for analysing eukaryotic proteins toward a deep understanding of their functions and interactions.
PREDICTION SCHEME AND CONTENTS
Overview
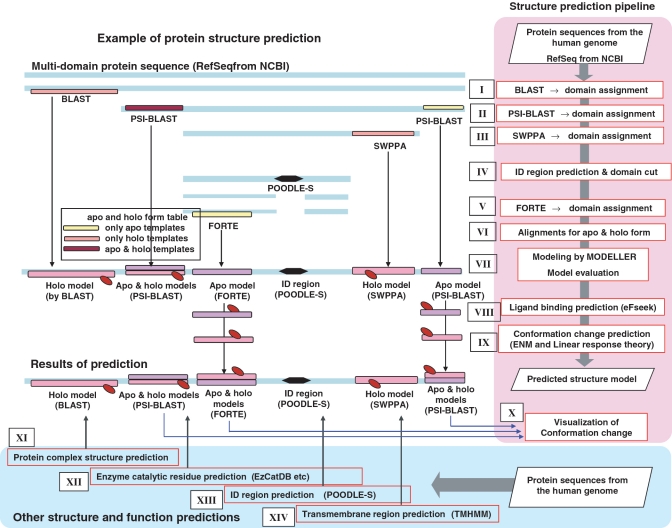
Schematically, two types of prediction systems were used to analyse protein sequences [RefSeq sequence (22)] automatically. One is the ‘Structure prediction pipeline’ (right pink regions in Figure 1) in which several homology search and protein structure prediction tools, conducting sequence–sequence, sequence–profile and profile–profile alignments, are combined sequentially, and it processes protein sequences, assigns them with 3D templates and finally produces 3D models. If available, 3D models of apo and holo forms were generated. The other components are ‘Other structure and function predictors’ (bottom light blue regions in Figure 1). They are an ensemble of independent prediction tools, which analyse protein sequences. All the results from these systems were accumulated in SAHG in XML formats.
Figure 1.
SAHG prediction systems. ‘Structure prediction pipeline’ and ‘Other structure and function predictions’ are shown in the right pink regions and bottom light-blue regions, respectively. The center panel illustrates each procedure in the flow of the structure prediction pipeline, showing how the results of systems are integrated. SWPPA: Smith–Waterman profile–profile alignment method; ID: intrinsically disordered; ENM: elastic network model.
Structure prediction pipeline
Construction of 3D models
Protein structure prediction consists of the following procedures: template searches and selection, alignment of target sequence and template, building 3D models and evaluation of model quality.
The template searches and their assignments to a target protein are the ‘step-wise-multi-methods’ approach. In the first step, a BLAST (23) search against all the latest Protein Data Bank (PDB) (7) and Structure Classification of Proteins (SCOP) (24,25) sequences is performed with 10−5 E-value cut-off. We selected templates, at least 90% of whose sequence could be aligned with the target, to ensure that the 3D models corresponded to stable domains or proteins. The resulting target sequence-template alignments were ranked based on their E-values. The best combination of templates for each domain was determined using an original algorithm to maximize the coverage of the target sequence (label I in Figure 1). In the second step, a PSI-BLAST (23) search with the same parameters was conducted for the remaining regions of the target sequence, where no models had been assigned and the best templates were assigned onto the target sequence (II in Figure 1). Protein sequence profiles were prepared using the latest NCBI-nr database. In the third step, a Smith–Waterman profile–profile alignment method (SWPPA) (26) was applied to the remaining regions against restricted templates (SCOP and PDB subsets with less than 40% sequence identity) with a cut-off of Z-score > 10, the comparable threshold to E-value < 10−5 in PSI-BLAST (III in Figure 1). Finally, the FORTE (27) search, a profile–profile comparison method, was performed for the remaining regions, with a strict cut-off of Z-score > 20, to detect distantly related templates (V in Figure 1). FORTE is based on the global–local alignment method and was adjusted to perform best (28) when the target proteins were almost the same length as the PDB entries (around 400 aa) (29). However, more than half of human proteins (53%) are larger than 400 amino acids and even the remaining regions are sometimes over 2000 amino acids. Thus, prior to the FORTE search, potential domains were carved out from the remaining regions using an algorithm based on the prediction of ID regions (IV in Figure 1) and fed into FORTE (see ‘Prediction of potential domains’ section for details).
Once the target sequence-template alignments were obtained, all templates were checked against our ‘apo and holo form table’ originally prepared by us (see ‘Apo and holo form table’ section in Supplementary data). For the template in apo form, the corresponding template (>90% sequence identity) in holo form was selected from the table and vice versa. For both the templates, alignments to target sequences were prepared (VI in Figure 1). In the model building and quality assessment step, 10 models were constructed using the MODELLER (30) software. The quality of the models was evaluated using Stability score (31) and the best 3D model for each alignment was chosen (VII in Figure 1).
As of July 2010, 24 878 RefSeq sequences [(22), 14 012 591 residues] encoded in the human genome were processed by the pipeline. In total, 42 581 structure models were constructed, of which 18 228, 14 577, 9163 and 613 templates were detected by BLAST, PSI-BLAST, SWPPA and FORTE, respectively. For 4083 models (9% of all models), both the apo and holo forms were assigned. In total, 35 275 residues were predicted to form long ID regions and removed from target sequences, in advance of the FORTE search. In total, 295 309 residues were eliminated because they were fragmented into small pieces (<26 residues). Multiple models were generated for 9057 RefSeq sequences, while only one model was generated for 12 310 RefSeq sequences. In total, 3511 RefSeq sequences remain without any predicted model. Note that one model does not necessarily correspond to one domain (sometimes it corresponds to a protein chain), but at least more than one-third of human proteins were estimated to be multi-domain proteins. In some cases, we assessed predictions by comparing models with the protein structures recently revealed. Even the sequence identities of the alignments are quite low (<20%), more than half predictions detect correct folds (Supplementary Table S1), indicating that our prediction pipeline worked well.
Treatments of multi domain proteins
Many human proteins are composed of multiple domains and contain a significant fraction of ID regions, as was described above. These factors often prevent predicting protein structures in their full-length forms. As a result, SAHG principally exhibits protein structure as an array of domains. However, when multi-domain structures are available in the templates, the prediction pipeline implicitly prioritizes them to take advantage of the relative domain orientations. The pool of templates consists of SCOP (24,25) domains and whole PDB (7) structures, some of which are not deposited in SCOP. At the template assignment step (I, II, III, V in Figure 1), a set of templates was chosen to maximize the length of modeled regions. This approach is effective in accepting PDB structures spanning multiple domains, as the templates.
Prediction of potential domains
ID regions were predicted using the POODLE-S (18) software, which calculates the probability of being in ID regions for each residue (XIII in Figure 1). As ID regions are considered to play fundamental roles in biological activities (17), their detections should be important. On the other hand, it is necessary to remove long ID regions from the target sequences and assign potential domain regions to assure better performance in structure prediction (FORTE search, V in Figure 1). For this purpose, we evaluated an existing method to predict domain boundaries [Domcut (32)] and found that it was likely to overcut potential domain regions into segments. For other methods (33–35), the same tendency was reported. We considered that the over-prediction was rather disadvantageous for arranging the input sequences for FORTE and developed a new method whose prediction was more ‘moderate’ (containing fewer false positives but more false negatives) based on the results of ID region prediction (IV in Figure 1), since ID regions act as linkers of structural domains (36). First, the results of POODLE-S for a target sequence were converted into a binary sequence in which 0 (P < 0.5) and 1 represent residues in structured regions and that in ID regions, respectively. Next, to detect regions where 0 were continuously abundant, we employed a simple two-state Hidden Markov Model. In this model, one state, ‘a mostly structured region’ (STR), emits 0 more frequently than 1 and the other state, ‘a mostly ID region’ (IDR), emits 1 more frequently than 0. The transition probability between STR and IDR and all the emission probabilities were empirically adjusted to eliminate over-prediction by referring to known domain data in PDB. Finally, the STR regions were estimated from the input binary sequence by calculating a Viterbi path.
Prediction of conformational change upon ligand binding
When templates for both the ligand-bound state (holo form) and unbound state (apo form) were detected using the ‘apo and holo form table’, two types of models were constructed and their structural changes upon ligand-binding are visualized by means of a morphing technique (the MORPH2 program in Martz-Authored PDB Tools see http://www.umass.edu/microbio/rasmol/pdbtools.htm) (X in Figure 1). The animation of conformational change provides significant information for protein function when it is shown with functional residues and ligands.
When there was only the template for apo form available and accordingly, only the model for apo form was constructed, its putative ligand and the binding sites were predicted by the eF-seek software (37) (VIII in Figure 1). eF-seek finds potential ligand-binding sites in the model of the apo form, if similar structures were deposited in eF-site, the database of representative ligand-binding sites (38). eF-seek employs a clique search algorithm. As this method is sensitive to the input 3D coordinates, the application was limited to the case of highly accurate structure models being available, i.e. the templates were detected by BLAST search with more than 90% sequence identity to the target sequences. The structural changes upon the predicted ligand-binding were then deduced using the elastic network model (39) and linear response theory to construct a model of the holo form (40) (IX in Figure 1).
Note that this approach and presentation is one of the key features of the SAHG database. Animated views of the conformational change of the domains upon ligand-binding could present a deep insight into the protein structure and function relationship (X in Figure 1). As of July 2010, conformational changes upon ligand-binding were predicted for 4083 modeled domains among 42 581 3D models.
Other structure and function predictors
Prediction of protein complex structure
In total, 33 687 protein complex structures were gathered from the PQS database (41). If all the subunits from two complexes were paired with more than 95% sequence identity, the complexes were clustered together in the single-linkage manner. The complex structure with the highest resolution was selected in each cluster of complexes and we obtained a non-redundant set composed of 12 730 template complexes. If a target sequence was related to a given subunit of a template complex with >80% sequence identity by the BLAST search and all the other subunits were related to any target sequences, the complex model was constructed by MODELLER. In total, 8667 complex models were prepared for 3650 target sequences (XI in Figure 1).
Ligand binding information
The ligands and their binding sites were retrieved from constructed models. The ligands were mainly small molecules, such as peptides, nucleotides, metal ions, etc. and some trivial chemicals from buffers or precipitants were excluded. Binding sites were residues whose distances from any ligand atoms were within 5 Å.
Prediction of catalytic residues
For the target sequences of enzymes, catalytic residues were predicted using the EzCatDB database (42) (XII in Figure 1). The EzCatDB database provides annotations on catalytic residues with PDB structure data. The catalytic residues and their positions were already denoted for sequences in the UniProt database (6), as mapped from the catalytic residues on the PDB sequence data, by BLAST search with 10−10 E-value cut-off and POA ver. 2.0 (43). From the human proteins in the UniProt database, target sequences were detected and catalytic residues were assigned in the same manner. Only chemically consistent residues were regarded as catalytic residues. The annotated ‘ACT_SITE’ residues for the human proteins in the UniProt database were also mapped on the target sequences using BLAST search.
Prediction of ID and transmembrane regions
ID regions were predicted by the POODLE-S software (XIII in Figure 1). Transmembrane regions were assigned by the TMHMM software (44) (XIV in Figure 1). If these predicted regions were overlapped with 3D models, the latter take priority over the former.
ACCESS AND INTERFACE
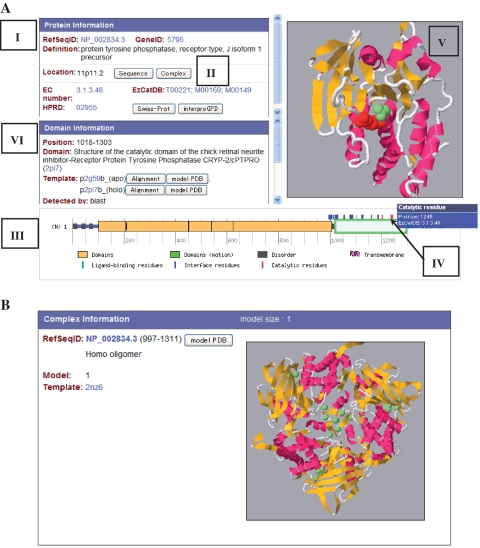
SAHG provides its graphical web interface at http://bird.cbrc.jp/sahg. By clicking a chromosome's image, all proteins coded in the chromosome are listed with the predicted models. By choosing an image of a domain, detailed information of the target protein is shown. More practically, detailed information of specific proteins can be accessed by querying with Gene ID, RefSeq ID, annotation keywords or their combinations or by sequence homology search (BLAST), from an ‘Advanced search page’. In the detailed information page (Figure 2A), all contents for a given protein are shown. The ‘Protein information’ panel provides the information of the protein's RefSeq ID (I in Figure 2A). The sequence in FASTA format is displayed by clicking a ‘Sequence’ button. Predicted protein complexes are shown via a ‘Complex’ button if available (II in Figure 2A). An example of a ‘complex information’ page is shown in Figure 2B. Links to EC number, EzCatDB (42), HPRD (45), Swiss-Prot(6) and InterPro (46) are provided if available. A bar indicator is convenient for seeing the position of the predicted models in the full-length protein (III in Figure 2A). It also shows the annotation of ligand-binding residues (retrieved from the holo models), protein–protein interface residues (from protein complexes), catalytic residues (from EzCatDB), ID regions (by POODLE-S) and transmembrane regions (by TMHMM). By pointing at the colored pins on the bar indicator with a mouse, precise locations (residue numbers) of ligand-binding residues (green pins), protein–protein interface residues (blue) or catalytic residues (red) are shown (see IV in Figure 2A, an example of a catalytic residue). When a modeled region in the bar indicator (blocks on the bar) is selected by clicking, the predicted 3D model appears in the Jmol window (an open-source Java viewer for chemical structures in 3D; see http://www.jmol.org/Jmol) (V in Figure 2A). When models of both apo and holo forms are available (green block on the bar), their structural changes upon ligand-binding are visualized by the morphing technique (the MORPH2 program in Martz-Authored PDB Tools; see http://www.umass.edu/microbio/rasmol/pdbtools.htm) and displayed as an animated image including the ligand molecules in this window. By clicking the bar indicator of ligand-binding or catalytic residues, the corresponding residues are highlighted in ‘CPK spacefill’ scheme in the Jmol window. The ‘Domain Information’ panel shows structural and functional information about a selected model (VI in Figure 2A). The target sequence-template alignments are displayed by an ‘Alignment button’. The predicted model can be downloaded in a pdb format via ‘model PDB’ button. Ligand-binding residues, protein–protein interface residues and catalytic residues are also listed as ‘Functional Residues’ in the same color of the bar indicator. (In Figure 2A, the ‘Domain information’ panel should be scrolled up).
Figure 2.
(A) Example view of SAHGs detailed information page [RefSeqID: NP_002834.3, protein tyrosine phosphatase, receptor type, J isoform 1 precursor (48)]. Labels I, II, III, IV, V and VI indicate the ‘Protein information’ panel, the ‘Complex’ button, the ‘bar indicator’, the ‘Domain information’ panel, the ‘Jmol Window’ and the ‘Catalytic residue’ pin on the bar indicator, respectively. (B) Example view of a ‘Complex information’ page (NP_002834.3). For this protein, only one complex structure in a homo-trimeric form was predicted.
FUTURE DIRECTIONS
To improve the accuracy of structure prediction we are implementing a probabilistic profile–profile alignment method in our prediction pipeline. The method is an enhanced version of the probabilistic sequence–sequence alignment method (47), which has been proven to perform better than PSI-BLAST, in particular for orphan proteins. New versions of structure models provided by the new pipeline will appear in fall of 2010. The results of predictions are being examined to clarify the function and the interaction of human proteins. For some proteins, predicted ligands are being verified experimentally. The structure model set in SAHG will be downloadable in bulk in future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Science and Technology Agency (JST) – Institute for Bioinformatics Research and Development (BIRD). Funding for open access charge: National Institute of Advanced Industrial Science and Technology (AIST).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Takatsugu Hirokawa and Kiyoshi Asai for their support of the project, to Martin Frith for his critical reading of the article and to Mari Saito for her contribution to website design.
REFERENCES
- 1.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 3.Zhang W, Dolan ME. Impact of the 1000 genomes project on the next wave of pharmacogenomic discovery. Pharmacogenomics. 2010;11:249–256. doi: 10.2217/pgs.09.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metzker ML. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 5.MacLean D, Jones JD, Studholme DJ. Application of ‘next-generation’ sequencing technologies to microbial genetics. Nat. Rev. Microbiol. 2009;7:287–296. doi: 10.1038/nrmicro2122. [DOI] [PubMed] [Google Scholar]
- 6.Consortium U. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie L, Bourne PE. Functional coverage of the human genome by existing structures, structural genomics targets, and homology models. PLoS Comput. Biol. 2005;1:e31. doi: 10.1371/journal.pcbi.0010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornton JM, Todd AE, Milburn D, Borkakoti N, Orengo CA. From structure to function: approaches and limitations. Nat. Struct. Biol. 2000;7(Suppl.):991–994. doi: 10.1038/80784. [DOI] [PubMed] [Google Scholar]
- 10.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77(Suppl. 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl. 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 12.Grant MA. Protein structure prediction in structure-based ligand design and virtual screening. Comb. Chem. High Throughput Screen. 2009;12:940–960. doi: 10.2174/138620709789824718. [DOI] [PubMed] [Google Scholar]
- 13.Katritch V, Rueda M, Lam PC, Yeager M, Abagyan R. GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex. Proteins. 2010;78:197–211. doi: 10.1002/prot.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl. 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 16.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 17.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu K, Muraoka Y, Hirose S, Tomii K, Noguchi T. Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics. 2007;8:78. doi: 10.1186/1471-2105-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, et al. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Dunbrack RL., Jr Scoring profile-to-profile sequence alignments. Protein Sci. 2004;13:1612–1626. doi: 10.1110/ps.03601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomii K, Akiyama Y. FORTE: a profile-profile comparison tool for protein fold recognition. Bioinformatics. 2004;20:594–595. doi: 10.1093/bioinformatics/btg474. [DOI] [PubMed] [Google Scholar]
- 28.Tomii K, Hirokawa T, Motono C. Protein structure prediction using a variety of profile libraries and 3D verification. Proteins. 2005;61(Suppl. 7):114–121. doi: 10.1002/prot.20727. [DOI] [PubMed] [Google Scholar]
- 29.Thornton JM, Orengo CA, Todd AE, Pearl FM. Protein folds, functions and evolution. J. Mol. Biol. 1999;293:333–342. doi: 10.1006/jmbi.1999.3054. [DOI] [PubMed] [Google Scholar]
- 30.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 31.Ota M, Isogai Y, Nishikawa K. Knowledge-based potential defined for a rotamer library to design protein sequences. Protein Eng. 2001;14:557–564. doi: 10.1093/protein/14.8.557. [DOI] [PubMed] [Google Scholar]
- 32.Suyama M, Ohara O. DomCut: prediction of inter-domain linker regions in amino acid sequences. Bioinformatics. 2003;19:673–674. doi: 10.1093/bioinformatics/btg031. [DOI] [PubMed] [Google Scholar]
- 33.Cheng J. DOMAC: an accurate, hybrid protein domain prediction server. Nucleic Acids Res. 2007;35:W354–W356. doi: 10.1093/nar/gkm390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebina T, Toh H, Kuroda Y. Loop-length-dependent SVM prediction of domain linkers for high-throughput structural proteomics. Biopolymers. 2009;92:1–8. doi: 10.1002/bip.21105. [DOI] [PubMed] [Google Scholar]
- 35.Kim DE, Chivian D, Malmstrom L, Baker D. Automated prediction of domain boundaries in CASP6 targets using Ginzu and RosettaDOM. Proteins. 2005;61(Suppl. 7):193–200. doi: 10.1002/prot.20737. [DOI] [PubMed] [Google Scholar]
- 36.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita K, Murakami Y, Nakamura H. eF-seek: prediction of the functional sites of proteins by searching for similar electrostatic potential and molecular surface shape. Nucleic Acids Res. 2007;35:W398–W402. doi: 10.1093/nar/gkm351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- 39.Tirion MM. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys. Rev. Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 40.Ikeguchi M, Ueno J, Sato M, Kidera A. Protein structural change upon ligand binding: linear response theory. Phys. Rev. Lett. 2005;94:078102. doi: 10.1103/PhysRevLett.94.078102. [DOI] [PubMed] [Google Scholar]
- 41.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 42.Nagano N. EzCatDB: the enzyme catalytic-mechanism database. Nucleic Acids Res. 2005;33:D407–D412. doi: 10.1093/nar/gki080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grasso C, Lee C. Combining partial order alignment and progressive multiple sequence alignment increases alignment speed and scalability to very large alignment problems. Bioinformatics. 2004;20:1546–1556. doi: 10.1093/bioinformatics/bth126. [DOI] [PubMed] [Google Scholar]
- 44.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 45.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koike R, Kinoshita K, Kidera A. Probabilistic alignment detects remote homology in a pair of protein sequences without homologous sequence information. Proteins. 2007;66:655–663. doi: 10.1002/prot.21240. [DOI] [PubMed] [Google Scholar]
- 48.Ostman A, Yang Q, Tonks NK. Expression of DEP-1, a receptor-like protein-tyrosine-phosphatase, is enhanced with increasing cell density. Proc. Natl Acad. Sci. USA. 1994;91:9680–9684. doi: 10.1073/pnas.91.21.9680. [DOI] [PMC free article] [PubMed] [Google Scholar]