Abstract
ArachnoServer (www.arachnoserver.org) is a manually curated database providing information on the sequence, structure and biological activity of protein toxins from spider venoms. These proteins are of interest to a wide range of biologists due to their diverse applications in medicine, neuroscience, pharmacology, drug discovery and agriculture. ArachnoServer currently manages 1078 protein sequences, 759 nucleic acid sequences and 56 protein structures. Key features of ArachnoServer include a molecular target ontology designed specifically for venom toxins, current and historic taxonomic information and a powerful advanced search interface. The following significant improvements have been implemented in version 2.0: (i) the average and monoisotopic molecular masses of both the reduced and oxidized form of each mature toxin are provided; (ii) the advanced search feature now enables searches on the basis of toxin mass, external database accession numbers and publication date in ArachnoServer; (iii) toxins can now be browsed on the basis of their phyletic specificity; (iv) rapid BLAST searches based on the mature toxin sequence can be performed directly from the toxin card; (v) private silos can be requested from research groups engaged in venoms-based research, enabling them to easily manage and securely store data during the process of toxin discovery; and (vi) a detailed user manual is now available.
INTRODUCTION
The growing realisation that most venomous animals possess a complex repertoire of protein toxins with potential pharmaceutical and agrochemical applications has led to an exponential increase in the rate of toxin discovery (1). Several databases have been specifically developed to facilitate retrieval of information about these toxins, such as Tox-Prot (2) and the Animal Toxin Database (ATDB) (3). These databases are critical for comparison of toxins across different groups of venomous animals but they typically lack the rich information content of manually curated databases that deal with specific subsets of animal toxins, such as ConoServer (4), which provides information about toxins from marine cone snails.
Spiders are the most evolutionarily successful venomous animals and the most abundant terrestrial predators. Their remarkable success is due in large part to their evolution of a pharmacologically complex venom that ensures rapid subjugation of prey. Most spider venoms are dominated by disulfide-rich peptide toxins that typically have high affinity and selectivity for specific subtypes of ion channels and receptors, making them particularly valuable from a pharmacological and drug discovery perspective (5–8). Spider venoms are likely to contain >10 million bioactive peptides based on the extraordinary taxonomic diversity of spiders, with the number of extant species predicted to be more than 100 000 and the demonstration that some venoms contain more than 1000 unique peptides (5). This is a larger pharmacopeia than that of all other venomous animals combined.
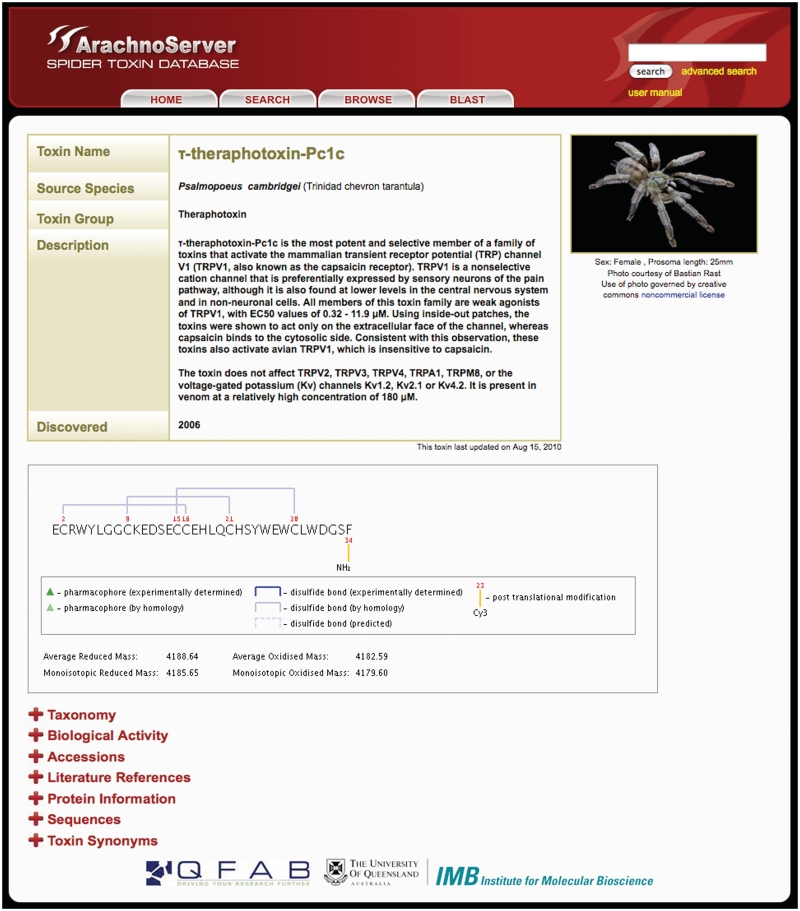
Prior to the introduction of ArachnoServer 1.0 (9), no publicly accessible database existed specifically for collating information about proteinaceous spider toxins. Since its establishment in 2009, ArachnoServer has been widely used and the website has hosted visitors from more than 60 countries. ArachnoServer toxin cards (see Figure 1 for an example) are now cross-referenced through links on the corresponding sequence records in UniProtKB (http://www.uniprot.org), ArachnoServer accession numbers are included in the UniProtKB mapping service and the UniProtKB records have adopted the rational toxin nomenclature (1) that has been applied universally in ArachnoServer.
Figure 1.
ArachnoServer Toxin Card for τ-theraphotoxin-Pc1c, a TRPV1 agonist isolated from the venom of the Trinidad chevron tarantula Psalmopoeus cambridgei. The top section of the Toxin Card provides a summary of the toxin’s activity, the source species and year of discovery. The dynamically generated image of the toxin’s primary structure displays the mature toxin sequence and, where available, its disulfide-bond framework, pharmacophore residues and posttranslational modifications. A new addition in Version 2.0 is the automated display of the average and monoisotopic masses for both the reduced and oxidized forms of the toxin. Below this image are additional user-expandable sections that provide information about the toxin’s molecular target, phyletic specificity and 3D structure, as well as literature references, toxin synonyms and both current and historic taxonomy of the source species. Clicking the spider photo yields a downloadable high-resolution image.
DATA SOURCES AND CURATION
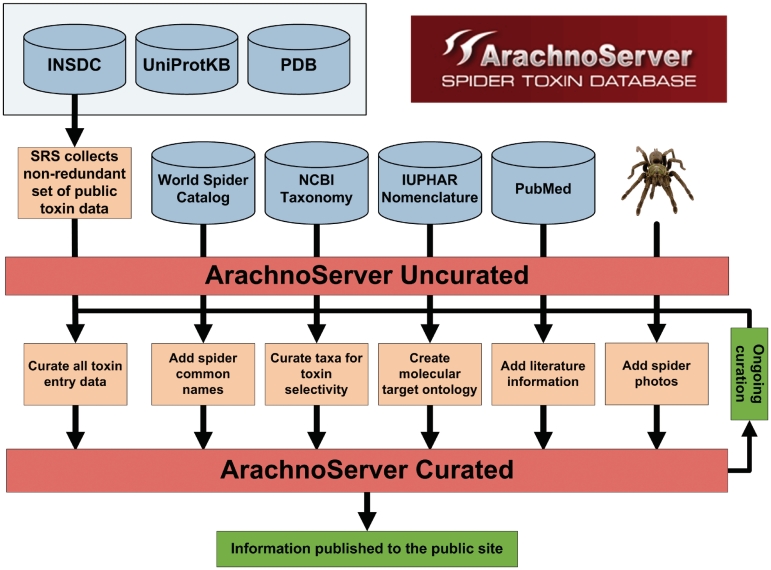
The starting point for data curation in ArachnoServer is the automated collection of all publically available sequence and annotation information for spider toxins from UniProtKB (10), the International Nucleotide Sequence Database Collection (INSDC) and the Protein Data Bank (PDB) (11). These data sets are joined with the assistance of the Sequence Retrieval System (SRS) (12) into a single non-redundant set containing peptide sequences, nucleotide sequences and protein structures (where available) for each toxin (Figure 2). Other database identifiers (e.g. NCBI taxonomy codes, Gene Ontology classifications, PROSITE and Pfam accessions, etc.) are also imported, as well as literature references, annotations of sequence and structure features (e.g. known locations of disulfide bonds) and toxin descriptions. Spider taxonomy is derived from the World Spider Catalog (13) while other taxonomy (used for classification of a toxin’s phyletic selectivity) is from the NCBI Taxonomy database (14). Since the majority of spider toxins act on ion channels and cell-surface receptors (8,15), we developed a molecular target ontology specifically for venom toxins that is based on the channel and receptor subtype definitions and nomenclature recommended by the International Union of Basic and Clinical Pharmacology (IUPHAR) (16).
Figure 2.
Schematic overview of the data retrieval and curation process in ArachnoServer.
Using the curation interface within ArachnoServer, curators can add additional data which includes, but is not limited to, a detailed description of the toxin; toxin name [which conforms to the rational nomenclature proposed for venom peptides (1) and venom sphingomyelinases (17)]; source species; discovery date; toxin synonyms; biological activity; phyletic specificity; molecular targets; sequence features such as toxin pharmacophore and disulfide bonds; database cross-references; and literature references. Literature references are sourced from PubMed (where available) using the PubMed eFetch web service. The process of data retrieval and curation in ArachnoServer is summarized in Figure 2.
IMPLEMENTATION
ArachnoServer is a Java Spring Model-View-Controller web application, built using a Hibernate Object Relational Model interfacing to a MySQL database. The web interface utilizes Asynchronous JavaScript and XML (AJAX) and dynamically generated HTML pages from server side JSP scripts to provide a rich application environment for both curation and public access. Care has been taken to ensure that the search, browse and BLAST features are powerful and intuitive. All major web browsers are supported.
IMPROVED FEATURES IN ARACHNOSERVER 2.0
In response to user feedback, we have implemented a major upgrade of ArachnoServer that includes the following new features:
Molecular mass
Mass spectrometry is becoming one of the standard methods used to characterize venom components (7). Most spider toxins contain multiple disulfide bonds and therefore the mass of the oxidized form of the peptide (as opposed to the reduced form that is calculated in most databases) is typically of primary interest to venom researchers. Thus, toxin records in ArachnoServer now provide the average and monoisotopic masses for both the reduced and oxidized form of the toxin (see Figure 1 for an example). Moreover, the advanced search feature now includes an option to perform a search based on any of these mass classes (see below).
Improved advanced search
ArachnoServer provides an Advanced Search feature that enables multiple search clauses to be grouped and joined using Boolean operators. We have now added the ability to search for toxins on the basis of toxin mass, external database accession numbers and the date on which the toxin record was published in ArachnoServer. Additional columns have been added to the table of search results that display context-specific data for certain search categories. For example, a search for toxins with oxidized molecular masses within a certain mass range will yield a table in which the oxidized molecular mass for each ‘hit’ is indicated in the additional column. Context-specific data columns have also been introduced for searches based on toxin discovery date as well as the number of solved PDB structures, number of biological activities, number of molecular targets, number of posttranslational modifications and number of disulfide bonds. Search results can be exported in both PDF and XML formats.
Additional BLAST capabilities
Rapid BLAST searches based on the mature toxin sequence ‘only’ can now be performed directly from the toxin card. This option provides a mechanism to enrich alignment results for mature toxin sequences. The option to perform a BLAST search using the entire toxin sequence, which may include signal and propeptide regions, is still available. BLAST results are formatted in HTML and contain links to ArachnoServer toxin cards and the corresponding UniProtKB records where available.
Browsing based on phyletic specificity
ArachnoServer includes a browse feature that initially enabled toxin records to be located on the basis of ‘Araneae Taxonomy, Molecular Targets and Posttranslational Modifications’. Each category creates a different browsing tree on the right hand side of the screen for easy selection of toxins. Toxins can now also be located on the basis of their ‘Phyletic Specificity’, that is, the range of organisms against which they are active. Choosing to browse by phyletic specificity creates a tree of organisms that includes five taxonomic levels: domain, class, order, genus and species. Selecting, for example the order Insecta will display all toxins with reported insecticidal activity. This new browse feature complements the ability to search for specific types of biological activity using the Advanced Search feature.
Private silos
We have created private silos, available upon request, for researchers actively involved in discovery and characterization of spider-venom toxins. These silos provide secure repositories for groups of researchers, enabling them to enter and manage their toxin sequences. Within a private silo, the nominated curators have access (via secure login) to the full suite of ArachnoServer curation tools and features, including the ability to securely BLAST their toxin sequences against the public ArachnoServer database. Toxin records within private silos remain strictly confidential until a researcher decides to release a toxin card to the ArachnoServer curators. We anticipate that private silos will not only help toxinologists manage their intellectual property but will also enhance the quality of ArachnoServer records by ensuring that the initial curation is done by the researchers who discovered the toxin.
User manual
A detailed user manual is now available for download from the ArachnoServer website. The manual details the kind of data stored in ArachnoServer and how it is curated. The user manual explains all of the features available within ArachnoServer and it provides examples of browsing, advanced searches and BLAST searches.
CURRENT STATUS OF ARACHNOSERVER
ArachnoServer 2.0 currently manages 1078 protein sequences, 759 nucleic acid sequences and 56 protein structures, an increase from 567, 334 and 51, respectively, in version 1.0. Overall, this represents the largest single collection of spider toxin records in any online database. In addition, version 2.0 contains many more high-resolution images of spiders from which toxins in the database have been sourced. All images are downloadable and are freely available for academic use according to the creative commons noncommercial license.
SUMMARY
ArachnoServer was designed to be useful to scientists across a broad range of disciplines, including pharmacologists, neuroscientists, medicinal chemists, toxinologists, structural biologists and clinicians. Version 2.0 of the database increases its utility by including many new features, a large increase in the number of curated protein and nucleic acid sequences and the most up-to-date information available on proteinaceous spider toxins.
FUNDING
Australian Research Council (Discovery Grants DP0774245 and DP0878450 to G.F.K.). Funding for open access charge: Australian Research Council.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors acknowledge financial support from the Australian Research Council, Dr Florence Jungo at the Swiss Institute of Bioinformatics for many helpful discussions and Bastian Rast for supplying high-resolution spider photographs.
REFERENCES
- 1.King GF, Gentz MC, Escoubas P, Nicholson GM. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon. 2008;52:264–276. doi: 10.1016/j.toxicon.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 2.Jungo F, Bairoch A. Tox-Prot, the toxin protein annotation program of the Swiss-Prot protein knowledgebase. Toxicon. 2005;45:293–301. doi: 10.1016/j.toxicon.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 3.He QY, He QZ, Deng XC, Yao L, Meng E, Liu ZH, Liang SP. ATDB: a uni-database platform for animal toxins. Nucleic Acids Res. 2008;36:D293–D297. doi: 10.1093/nar/gkm832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaas Q, Westermann JC, Halai R, Wang CK, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008;24:445–446. doi: 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- 5.Escoubas P, Sollod BL, King GF. Venom landscapes: mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47:650–663. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Estrada G, Villegas E, Corzo G. Spider venoms: a rich source of acylpolyamines and peptides as new leads for CNS drugs. Nat. Prod. Rep. 2007;24:145–161. doi: 10.1039/b603083c. [DOI] [PubMed] [Google Scholar]
- 7.Escoubas P, Quinton L, Nicholson GM. Venomics: unravelling the complexity of animal venoms with mass spectrometry. J. Mass Spectrom. 2008;43:279–295. doi: 10.1002/jms.1389. [DOI] [PubMed] [Google Scholar]
- 8.Vassilevski AA, Kozlov SA, Grishin EV. Molecular diversity of spider venom. Biochemistry. 2009;74:1505–1534. doi: 10.1134/s0006297909130069. [DOI] [PubMed] [Google Scholar]
- 9.Wood DL, Miljenovic T, Cai S, Raven RJ, Kaas Q, Escoubas P, Herzig V, Wilson D, King GF. ArachnoServer: a database of protein toxins from spiders. BMC Genomics. 2009;10:375. doi: 10.1186/1471-2164-10-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etzold T, Harris H, Beulah S. In: Bioinformatics: managing scientific data. Lacroix Z, Critchlow T, editors. San Francisco: Morgan Kaufmann; 2003. pp. 109–145. [Google Scholar]
- 13.Platnick NI. Advances in spider taxonomy, 1992–1995: with redescriptions 1940–1980 (updated online version available at http://research.amnh.org/entomology/spiders/catalog/) New York: New York Entomological Society & The American Museum of Natural History; 1997. [Google Scholar]
- 14.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sollod BL, Wilson D, Zhaxybayeva O, Gogarten JP, Drinkwater R, King GF. Were arachnids the first to use combinatorial peptide libraries? Peptides. 2005;26:131–139. doi: 10.1016/j.peptides.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Alexander SPH, Mathie A, Peters JA. Guide to Receptors and Channels (GRAC), 4th edn. Br. J. Pharmacol. 2009;158(Suppl. 1):S1–S254. doi: 10.1111/j.1476-5381.2009.00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binford GJ, Bodner MR, Cordes MH, Baldwin KL, Rynerson MR, Burns SN, Zobel-Thropp PA. Molecular evolution, functional variation, and proposed nomenclature of the gene family that includes sphingomyelinase D in sicariid spider venoms. Mol. Biol. Evol. 2009;26:547–566. doi: 10.1093/molbev/msn274. [DOI] [PMC free article] [PubMed] [Google Scholar]