Abstract
PREX (http://www.csb.wfu.edu/prex/) is a database of currently 3516 peroxiredoxin (Prx or PRDX) protein sequences unambiguously classified into one of six distinct subfamilies. Peroxiredoxins are a diverse and ubiquitous family of highly expressed, cysteine-dependent peroxidases that are important for antioxidant defense and for the regulation of cell signaling pathways in eukaryotes. Subfamily members were identified using the Deacon Active Site Profiler (DASP) bioinformatics tool to focus in on functionally relevant sequence fragments surrounding key residues required for protein activity. Searches of this database can be conducted by protein annotation, accession number, PDB ID, organism name or protein sequence. Output includes the subfamily to which each classified Prx belongs, accession and GI numbers, genus and species and the functional site signature used for classification. The query sequence is also presented aligned with a select group of Prxs for manual evaluation and interpretation by the user. A synopsis of the characteristics of members of each subfamily is also provided along with pertinent references.
INTRODUCTION
Peroxiredoxin (Prx or PRDX) proteins (EC 1.11.1.15) are a ubiquitous family of highly expressed, thioredoxin-scaffold enzymes that exhibit cysteine-dependent peroxidase activity with hydrogen peroxide and larger hydroperoxide substrates (1–3). These antioxidant enzymes are important in many organisms, including plants and bacteria as well as animals, for protecting against oxidative damage. Moreover, eukaryotic Prxs are implicated in intracellular signaling and the regulation of such processes as cell proliferation, differentiation and cell death (4). Prxs have also been shown to be induced by oxidative stress, to be aberrantly expressed in cancer and to impact the radiation sensitivity of cells (5). The importance of these proteins as potential prognostic or therapeutic targets is of widespread interest across many diseases that involve reactive oxygen species, including cancer and cardiovascular disease (6).
With the rapid increase over the last decade in the functional and structural information available about Prxs, distinctions between various Prx subfamilies have become apparent (e.g. oligomerization properties, location of mechanistically important cysteinyl residues). Prx proteins are widely distributed and members of multiple subfamilies are present in most organisms, including three subfamilies in humans. Prxs have frequently been classified based on the number of Cys residues involved in the catalytic cycle; however, the distinction between 2-Cys and 1-Cys function is not particularly useful as a global classifier because representatives of each type seem to exist within all the subfamilies (7). Bioinformatics efforts by others have helped clarify some of the features distinguishing Prxs subfamilies (3,8) and are particularly powerful when combined with structural analyses of representative Prxs (9,10). While structure-based subfamily classifications are increasingly recognized, detailed annotation to the level of Prx subfamily across the entire GenBank database remains scant and frequently confusing.
Only one other database is available where information for peroxidases has been collected and organized. PeroxiBase (http://peroxibase.toulouse.inra.fr/) (11) is a collection of sequences classified across all types of heme and non-heme peroxidases, which comprise the enzyme class EC 1.11.1.x. In contrast, PREX is focused on the detailed annotation of the Prxs, one type of non-heme peroxidase found in PeroxiBase. PeroxiBase organizes the Prxs into subfamilies based on broad biological function and overall sequence comparison (typical 1-Cys, typical 2-Cys, atypical 2-Cys and two different thioredoxin peroxidase subfamilies) (11). The information in PREX classifies the Prxs based on structural and sequence information at the reactive cysteine active site, resulting in six mechanistically based subfamilies. As of October 2010, PeroxiBase contains 826 proteins annotated as Prx, while PREX contains 3516 proteins that are each assigned to a single subfamily. Thus, the PREX information is complementary to the broader biologic functional organization provided in PeroxiBase.
To provide global Prx subfamily assignments for the Prx field, we have recently identified 3516 members of the Prx family (12) from the January 2008 GenBank database. Based upon structural analysis by Hall et al. (7) and by our own bioinformatic analysis (12) utilizing the functional site profiling method (also referred to as active site profiling) (13) and the functional site profile search tool implemented in Deacon Active Site Profiler (DASP) (14) to analyze sequence conservation in the structural vicinity of the catalytic cysteine, all of these proteins have been unambiguously classified into six functionally relevant subfamilies: AhpC/Prx1, Prx6, BCP/PrxQ, Tpx, Prx5 and AhpE (Table 1). We have incorporated our newly developed ‘index’ of proteins into the present searchable database, called PREX (http://www.csb.wfu.edu/prex/). PREX is designed to help fill the need for accurate classification of the Prx family members and to be useful for researchers familiar with Prx function, as well as for those new to the subject.
Table 1.
Summary of the Prx subfamilies present in PREX
| Subfamily | Number of database members | Canonical subfamily members | Phylogenetic distribution | Typical location of CR when presenta |
|---|---|---|---|---|
| AhpC/Prx1b | 1059 | Salmonella typhimurium AhpC, Homo sapiens Prx1 through Prx4 | Archea, Bacteria, Plants, Unicellular and Multicellular Eukaryotes | C-terminus (>96%)c |
| BCP/PrxQ | 1115 | Escherichia coli bacterioferritin comigratory protein, plant PrxQ | Bacteria, Plants | Helix α2 (∼50%) or α3 (∼7%)d |
| Prx5e | 517 | H. sapiens Prx5 | Bacteria, Eukaryotes | Helix α5 (∼17%)d |
| Prx6f | 493 | H. sapiens Prx6 | Archea, Bacteria, Plants, Unicellular and Multicellular Eukaryotes | No CR |
| Tpxg | 307 | E. coli Tpx | Bacteria | Helix α3 (>96%)d |
| AhpE | 25 | Mycobacterium tuberculosis AhpE | Bacteria | Unknownh |
aStructural designations as in (10). If no CR is present, resolving thiol must come from another protein or small molecule.
bThe AhpC/Prx1 subfamily is also known as the ‘typical 2-Cys’ Prxs and includes tryparedoxin peroxidases, Arabidopsis thaliana 2-Cys Prx, barley Bas1 and Saccharomyces cerevisiae TSA1 and TSA2.
cThe CR is near the C-terminus of the partner subunit within the homodimer; upon oxidation, intersubunit disulfide forms between the CP and the CR of the two chains.
dIntrasubunit disulfide formed in oxidized protein (so-called ‘atypical’ 2-Cys Prxs).
eThe Prx5 subfamily includes Populus trichocarpa PrxD, the plant type II Prxs, mammalian Prx5 and a group of bacterial glutaredoxin-Prx5 fusion proteins.
fThe Prx6 subfamily (frequently referred to as the ‘1-Cys’ group) also includes the bacterial Prx6 proteins, A. thaliana 1-Cys Prx and S. cerevisiae mitochondrial Prx1.
gThe Tpx subfamily includes bacterial proteins (e.g. from Streptococcus pneumoniae and Helicobacter pylori) named thiol peroxidase, p20 and scavengase.
hCanonical member contains no CR, but >50% of sequences include a potential CR in α2, similar to E. coli BCP.
DATABASE CONTENT
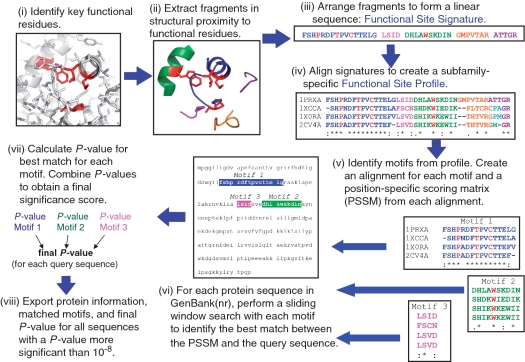
Prx classifications were made using the DASP profile search tool (14) (publically available at http://dasp.deac.wfu.edu/) that uses fine structure mapping and profiling to identify motifs of functional importance (13,15). DASP requires the selection of key residues that define a functional site within a protein family and then identifies nearby sequence fragments (Figure 1). As described in more detail in a separate research paper (12), key residues used to define the Prx active site included the three residues in the PXXX(T/S)XXC motif found in all Prxs (1,3,16) as well as a conserved Trp /Phe residue located ∼6 Å from the catalytic cysteine (Trp81 in Salmonella typhimurium AhpC). All residues which contained an atom located within 10 Å of the center of geometry of at least one of these key residues were extracted and the sequence fragments containing these residues were placed in order from N- to C-terminus to form the ‘functional site signature’ from all 29 non-redundant Prxs of known structure in the RCSB PDB database as of January 2008 (Figure 1).
Figure 1.
Identification of Prx sequences using the DASP tool. (i) The active site of human Prx6 (PDB identifier 1prx) is shown with the four key residues highlighted in red. (ii) Structural segments located within 10 Å of the center of geometry of the key catalytic residues are identified (each segment shown in a different color) and extracted from the global structure. (iii) The sequence fragments are then combined to form a functional site signature [residue colors correspond to the color of structure segments in (ii); key residues are highlighted in red]. (iv) Functional-site signatures for structurally characterized members of the Prx6 subfamily are aligned using ClustalW (22,24) to create a functional site profile. (v) Motifs are identified within any fragments that contain at least three residues and position specific scoring matrices (PSSM) (25) are created for each motif. (vi) For each sequence in a user-selected sequence database, the PSSM for each motif is used to find and score the segment within a query sequence which best matches a motif. (vii) Each time a motif is matched to a position in the protein sequence, a P-value is calculated that represents the probability of finding a match as good as the observed match within a random sequence. The P-values for all motifs in a single sequence are then combined using QFAST to obtain the final statistical significance score (final P-value) (26). (viii) The protein information (including accession numbers, annotations and species), final P-value and sequence fragments matched to each queried motif are exported for all sequences with a final P-value more significant than a user-selected P-value. See (13–15) for a more detailed description of DASP utilities and architecture.
Each Prx signature was assigned to a single subfamily (AhpC/Prx1, Prx6, BCP/PrxQ, Prx5, Tpx or AhpE) based on previously published structural characterizations (10) which agreed well with hierarchical clustering of the aligned functional site signatures. Signatures for multiple representatives in each subfamily were combined to generate a subfamily-specific ‘functional site profile’ that was used to identify subfamily members from GenBank(nr), according to the method described elsewhere (12,14). Each returned sequence is associated with a score (P-value) based on the probability of finding as good of a random match in a random sequence as the observed match. A P-value cutoff of 1 × 10−8 was selected to define ‘true’ DASP hits for each subfamily, as described elsewhere (12). Due to the limited number of structures available for the BCP/PrxQ and AhpE subfamilies, ‘engineered’ profiles (including sequence fragments and manually generated ‘signatures’ from structurally uncharacterized proteins) were created in order to increase the robustness of the profiles. The results for each subfamily were hand curated to remove any sequences that did not contain the PXXX(T/S)XXC Prx motif or that were identified with a more significant P-value in another Prx subfamily (62 sequences were removed out 3578 sequences originally identified for all subfamilies). Complete details are described in Nelson et al. (12).
DATABASE ARCHITECTURE
This subfamily-specific list of Prxs was used to create PREX, a relational database built on MySQL and accessed from a PHP web-based interface (http://www.csb.wfu.edu/prex/). The basic relationship diagram is provided in Supplementary Figure S1. The PREX database contains 3516 unique signatures representing 8658 GenBank identification numbers classified into only one of six Prx subfamilies. Each signature in the PREX database is associated with its GenBank annotations, organism of origin, the DASP subfamily assignment, and the full sequence of the protein from which it comes. To construct the local sequence database for BLAST searching, the full protein sequence associated with each signature was downloaded from GenBank and incorporated into a local BLAST + 2.2.23 database (17,18).
DATABASE INTERFACE ORGANIZATION
PREX has been designed to aid in the characterization of Prx proteins by providing a user friendly tool to identify the subfamily assignment for a Prx protein of interest and to quickly and easily find distinguishing features and typical characteristics for each Prx subfamily. Access and interface to the PREX database is provided by the publically available website, at http://www.csb.wfu.edu/prex/. This website is divided into three sections: (i) Peroxiredoxin Information, (ii) Search Tools and (iii) References.
Peroxiredoxin information
A general introduction to the Prx family is provided on the Home Page along with the list of subfamilies present in this database. Additional information for each subfamily can be accessed from the Prx Subfamily tab and provides structural and functional characteristics of members of each subfamily, PDB identifiers for structurally characterized subfamily members, and some pertinent references. In addition, a representative multiple sequence alignment is provided containing the functional site signatures for 4–5 members of the selected protein subfamily and one representative from each of the other subfamilies.
Search tools
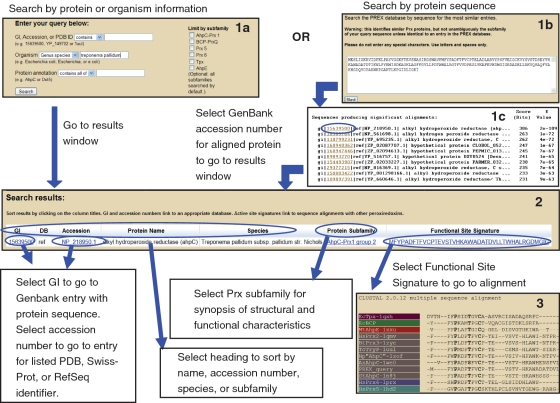
In order to identify the subfamily assignment of a particular Prx protein, searches of this database can be conducted by accession number, PDB ID, protein annotation, organism name or protein sequence (Figure 2). It should be noted that the BLAST search algorithm utilized for the sequence search relies on conservation across the entire protein sequence while subfamily assignments in this database are based exclusively on sequence conservation around the functional site using the DASP signatures. We therefore stress that caution must be exercised with any query sequence that is not an identical match with a pre-classified member of the PREX database. A stringent E-value of 1 × e−40 has been selected as the default cutoff based upon trial runs with both true Prx proteins and closely related decoy proteins.
Figure 2.
Examples of queries and results from PREX. The screenshot shown in Box 1a represents a taxonomy search for database members from Treponema pallidum. Text searches of the PREX database can also be conducted by GI, accession numbers, PDB ID or protein annotation. If a matching protein is identified, the user is taken directly to the results window; the screenshot shown in Box 2 represents the single Prx found in T. pallidum. Protein sequence searches of PREX (Box 1b) utilize BLAST to identify PREX database members with high sequence similarity to the query sequence. Shown in Box 1c is the BLAST output obtained after searching the full sequence of T. pallidum AhpC. Selecting the GI number of one of the identified proteins will direct the user to the results window for that PREX database member (Box 2). Selecting the functional site signature generates a multiple sequence alignment (Box 3) containing the functional site signatures for the selected PREX database protein (labeled as PREX_query), 4–5 selected members of the same subfamily and one representative from each of the other subfamilies. If accessed through a BLAST search, the multiple sequence alignment also includes the full sequence of the original sequence query (labeled as BLAST_query). Colors in Box 3 identify the subfamily assignment for each signature. In bold is the PXXX(T/S)XXC sequence motif that is invariant at the active site of Prx proteins (16).
Search output
Output for the text searches includes: identifying numbers (including Genbank accession numbers, PDB identifier and Swiss-Prot entry names), any associated annotations, genus and species, the subfamily to which the query sequence belongs and the functional site signature generated by DASP (Figure 2). This information is also accessible from the BLAST output screen for each PREX database protein identified as similar to a query sequence (by selecting the GI number). As described above, additional information for the relevant subfamily can be accessed through a link associated with the subfamily name. The GenBank entry containing the protein sequence is accessed through a link associated with the GI number and associated entries in UniProt (19), EMBL (20) and the RCSB Protein databank (21) may be accessed through links in the column labeled Accession. The output is in a format than can be easily sorted by the user.
In addition, a sequence alignment containing the functional site signature for each PREX entry aligned with signatures from 4 to 5 members of the same subfamily and a representative member from each of the other subfamilies can be accessed by selecting the functional site signature on the results page (Figure 2). Alignments are created with a local copy of ClustalW, version 2.0.12 (22) (parameters for Gap Open Penalty and Gap Extension Penalty are set to 5 and 0.5, respectively; all other parameters are set to default values). When a BLAST search has been performed, the alignment includes the full sequence of the protein query in addition to the chosen PREX database signature. Protein names are colored according to their subfamily assignments for all but the BLAST query sequence. This allows the user to verify the presence of the PXXXT/SXXC motif (this motif is defined as mandatory for a functional Prx) and compare the aligned query sequence with the functional site signatures of the selected PREX database member and other Prxs from within and outside that subfamily.
References
Resources pertaining to the Prx family are listed in the reference section, providing key links and literature references to help direct researchers new to the Prx field.
FURTHER DEVELOPMENTS AND DATABASE MAINTENANCE
In order to provide more flexible search options, functionality will be added to PREX by the implementation of a taxonomy browser that will allow the user to identify all Prxs within a user-selected taxonomic group. We also plan to develop a search algorithm based on DASP that will utilize subfamily profiles to evaluate subfamily assignment for user entered sequences. Because this method would rely on sequence conservation in the region of the functional site, it would allow users to obtain reliable subfamily assignments for proteins not currently found in the PREX database. Areas of future development are indicated by the blue ovals in Supplementary Figure S1.
Database maintenance and updating is crucial. As structures of additional Prx proteins become available, their signatures can be used to improve our subfamily profiles and, thus, our ability to identify and classify Prxs from the GenBank (nr) database (23). New searches of GenBank (nr) will provide annotations of newly deposited sequences. Because the current DASP search method is manual and somewhat laborious, we have established a collaboration which aims to develop an automated pipeline for doing this search. Once this pipeline is developed, we plan to identify new Prx structures from the RCSB protein databank (21) and to utilize the pipeline to produce profiles and search GenBank(nr) to generate an updated database regularly. References and subfamily descriptions will be updated yearly to include new key references and to describe current advances in the Prx field.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (MCB0517343 to J.S.F.); National Institutes of Health (F32 GM074537 to K.J.N. and RO1 GM050389 to L.B.P.). Calculations to identify and classify subfamily members for the database were performed on Wake Forest University's DEAC cluster, http://www.deac.wfu.edu, supported by a Shared University Research award from IBM, Inc. for storage hardware and by the Wake Forest Information Systems department. Funding for open access charge: MCB0517343 and F32 GM074537.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Special thanks to Stacy Reeves, Chananat Klomsiri, Derek Parsonage and Edward Pryor for advice and for database testing.
REFERENCES
- 1.Poole LB. The catalytic mechanism of peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. NY: Springer; 2007. pp. 61–81. [Google Scholar]
- 2.Wood ZA, Schröder E, Harris JR, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol. Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 4.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Wang Y, Su Y. Peroxiredoxins, a novel target in cancer radiotherapy. Cancer Lett. 2009;286:154–160. doi: 10.1016/j.canlet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- 6.Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol. Med. 2005;11:571–578. doi: 10.1016/j.molmed.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall A, Nelson K, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2010 doi: 10.1089/ars.2010.3624. doi:10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. NY: Springer; 2007. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- 9.Copley SD, Novak WR, Babbitt PC. Divergence of function in the thioredoxin fold suprafamily: evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–13995. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- 10.Karplus PA, Hall A. Structural survey of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. NY: Springer; 2007. pp. 41–60. [DOI] [PubMed] [Google Scholar]
- 11.Koua D, Cerutti L, Falquet L, Sigrist CJ, Theiler G, Hulo N, Dunand C. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 2009;37:D261–D266. doi: 10.1093/nar/gkn680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson KJ, Knutson ST, Soito L, Klomsiri C, Poole LB, Fetrow JS. Analysis of the peroxiredoxin family: using active site structure and sequence information for global classification and residue analysis. Proteins. 2010 doi: 10.1002/prot.22936. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cammer SA, Hoffman BT, Speir JA, Canady MA, Nelson MR, Knutson S, Gallina M, Baxter SM, Fetrow JS. Structure-based active site profiles for genome analysis and functional family subclassification. J. Mol. Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- 14.Huff RG, Bayram E, Tan H, Knutson ST, Knaggs MH, Richon AB, Santago P, 2nd, Fetrow JS. Chemical and structural diversity in cyclooxygenase protein active sites. Chem. Biodivers. 2005;2:1533–1552. doi: 10.1002/cbdv.200590125. [DOI] [PubMed] [Google Scholar]
- 15.Fetrow JS. Active site profiling to identify protein functional sites in sequences and structures using the Deacon Active Site Profiler (DASP) Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0810s14. Chapter 8, Unit 8 10. [DOI] [PubMed] [Google Scholar]
- 16.Fomenko DE, Gladyshev VN. Identity and functions of CxxC-derived motifs. Biochemistry. 2003;42:11214–11225. doi: 10.1021/bi034459s. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leinonen R, Akhtar R, Birney E, Bonfield J, Bower L, Corbett M, Cheng Y, Demiralp F, Faruque N, Goodgame N, et al. Improvements to services at the European Nucleotide Archive. Nucleic Acids Res. 2010;38:D39–D45. doi: 10.1093/nar/gkp998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 23.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey TL, Gribskov M. Methods and statistics for combining motif match scores. J. Comput. Biol. 1998;5:211–221. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- 26.Bailey TL, Gribskov M. Combining evidence using P-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]