Abstract
This article introduces the second release of the Gypsy Database of Mobile Genetic Elements (GyDB 2.0): a research project devoted to the evolutionary dynamics of viruses and transposable elements based on their phylogenetic classification (per lineage and protein domain). The Gypsy Database (GyDB) is a long-term project that is continuously progressing, and that owing to the high molecular diversity of mobile elements requires to be completed in several stages. GyDB 2.0 has been powered with a wiki to allow other researchers participate in the project. The current database stage and scope are long terminal repeats (LTR) retroelements and relatives. GyDB 2.0 is an update based on the analysis of Ty3/Gypsy, Retroviridae, Ty1/Copia and Bel/Pao LTR retroelements and the Caulimoviridae pararetroviruses of plants. Among other features, in terms of the aforementioned topics, this update adds: (i) a variety of descriptions and reviews distributed in multiple web pages; (ii) protein-based phylogenies, where phylogenetic levels are assigned to distinct classified elements; (iii) a collection of multiple alignments, lineage-specific hidden Markov models and consensus sequences, called GyDB collection; (iv) updated RefSeq databases and BLAST and HMM servers to facilitate sequence characterization of new LTR retroelement and caulimovirus queries; and (v) a bibliographic server. GyDB 2.0 is available at http://gydb.org.
INTRODUCTION
Mobile genetic elements (MGEs) are ubiquitous, autonomous genetic units that often constitute a significant part of their host genomes. It is commonly accepted that mobile DNA elements are powerful vectors for disease and evolution, from which distinct host genes have evolved during the history of life (1,2). The emergence and subsequent role played by viruses and MGEs in the history of life is an exciting topic that requires further investigation. In this respect, researchers aim to discern relevant aspects of the molecular changes responsible for various characteristics in organisms related to horizontal transfer, infection and disease. Among the distinct initiatives launched with the aim of investigating the diversity of MGEs (see for example 3–5) was the Gypsy Database (GyDB) of MGEs (6), a research project devoted to the evolutionary dynamics of viruses and MGEs (and their related host proteins), which was launched in 2008. The GyDB project is a highly informative database established within an evolutionary context of classification, where one piece of research delivers one conclusion that drives individuals towards another goal. The most captivating aspect of this project is that a share of our efforts are dedicated to the interpretation of analyses, paying particular attention to non-redundant elements displaying a certain degree of distance and investigating how they can be collectively aligned or related, in terms of protein domain architecture, with other lineages and elements. Because of the impressive molecular diversity of viruses and MGEs, the GyDB is a long-term project that has been arranged in a database in continuous progression, and must be achieved in stages. The current database stage and scope is retroviruses and retrotransposons with long terminal repeats (LTR retroelements) and their relatives. Following the outline of the earlier release (the study of Ty3/Gypsy and Retroviridae LTR retroelements), this article presents the GyDB update based on the phylogenetic evaluation of the most representative LTR retroelement families and the plant caulimoviruses. This update, called GyDB 2.0, is available at http://gydb.org and includes sequence phylogenetic classification in addition to significant bioinformatic improvements. In particular, the new infrastructure implements a wiki management system constructed with the aim of promoting a world-wide community of researchers collaborating in the analysis and classification of MGEs and viruses inhabiting (or circulating in) living organisms.
THE UPDATE: NEW FEATURES
GyDB 2.0 consists of 1234 web pages addressing the phylogenetic study of Ty3/Gypsy, Retroviridae, Ty1/Copia and Bel/Pao LTR retroelement. Caulimoviruses (Caulimoviridae) are formally plant DNA pararetroviruses, but they were considered in GyDB 2.0 owing to their relationship with LTR retroelements based on the common gag/coat and pol regions [for more details, see (7) and references therein]. Table 1 summarizes the topics addressed in this update, as well as the servers and database sections it offers. The sequences on which GyDB 2.0 is based were retrieved from GenBank (8) and the methodologies employed were the same as those described earlier in references (6,7,9). At GyDB we evaluate the phylogenetic signal of classified distinct elements and create hidden Markov model (HMMs) profiles (10) per lineage and protein domain. In addition, the project is concerned with the evolutionary relationships between MGEs and their host genomes, based on the analysis of common protein families. In this regard, GyDB 2.0 focuses on two protein superfamilies including protein products commonly encoded by LTR retroelements and their host genomes; the chromodomain superfamily (11) and clan AA of aspartic peptidases (12,13). This second release is accompanied by bibliographic data-mining from PubMed databases hosted at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) to document up to date information regarding the distinct classified elements.
Table 1.
GyDB 2.0 new features: topics and contents
| Systems | Families | Lineages | Elements | Protein domains | Accessory proteins | LTRs |
|---|---|---|---|---|---|---|
| LTR retroelements | Ty3/Gypsy | 34 | 96 | 8 | 1 | Yes |
| LTR retroelements | Ty1/Copia | 19 | 69 | 8 | – | Yes |
| LTR retroelements | Retroviridae | 8 | 50 | 8 | 41 | Yes |
| LTR retroelements | Bel/Pao | 5 | 23 | 7 | – | Yes |
| LTR retroelementsa | Caulimoviridae | 6 | 30 | 10 | 27 | No |
| Related families | Clan AA | 35 | 323 | 1 | – | No |
| Related families | Chromodomains | 2 | 123 | 1 | – | No |
| Topics | Sections | Availability |
|---|---|---|
| Systematics | 9 | Side menu |
| Domains | 14 | Side menu |
| Database | 8 | Side menu |
| Servers | 3 | Top menu: BLAST, HMM, Literature |
| Wiki tools and utilities | 3 | Top menu |
| Databases | Items | Sections | |
|---|---|---|---|
| Genomes (full-length genomes) | 271 | sequences | BLAST search and RefSeq DBs |
| LTRs (nucleotide sequences) | 413 | sequences | BLAST search and RefSeq DBs |
| Cores (protein cores sequences) | 1895 | sequences | BLAST search and RefSeq DBs |
| HMMs | 314 | HMM profiles | HMM search and GyDB collection |
| Multiple alignments | 131 | alignments | GyDB collection |
| Consensus sequences | 314 | MRC sequences | GyDB collection |
| Phylogenetic trees | 70 | trees | Phylogenies |
| Clan AA ancestral reconstruction | 70 | alignments | CAARD database |
| Literature | 100797 | references | Literature server |
aWe included caulimoviruses in the second release in view of their relationship with LTR retroelements based on the common gag/coat and pol region.
DATABASE ORGANIZATION
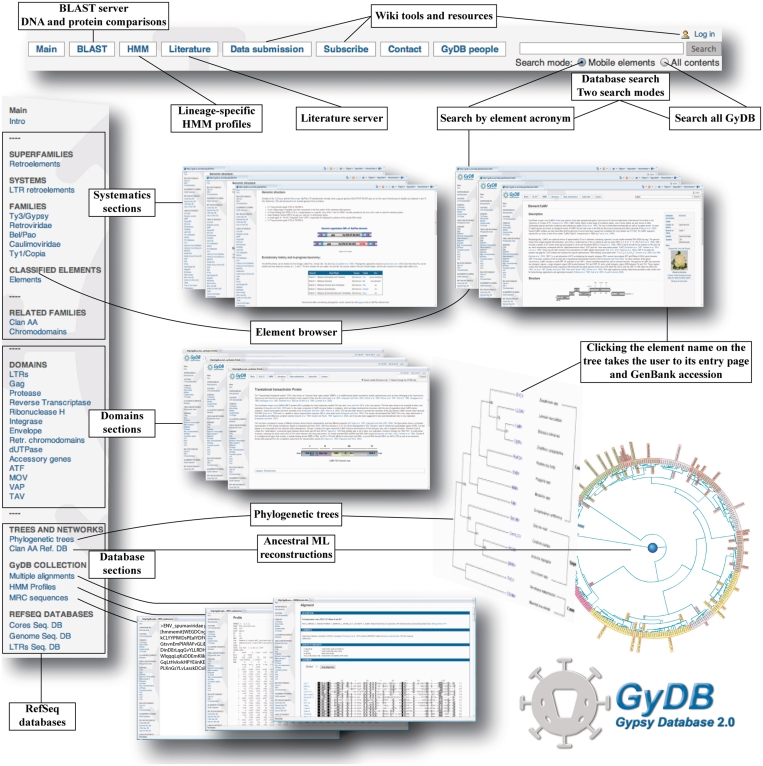
GyDB 2.0 is deployed over a Linux-MySQL-Apache-PHP (LAMP) stack, with additional Ajax programming to minimize server responses to client browsers. The design is similar to that of the previous release but implements various changes on the web interface. As shown in Figure 1, the database organization is founded upon two major menus––a top menu and a side menu. The top menu allows access to the three servers:
BLAST server; implements a BLAST search powered by the NCBI BLAST package (14), allowing protein and DNA comparisons with the GENOMES, LTRs and CORES databases. These databases collect the full-length genomes, the LTR sequences and all the protein sequences on which the second release is based, respectively.
HMM server; implements HMMER3 package (http://hmmer.janelia.org) and allows protein comparisons against a database of protein domain lineage-specific HMM profiles created based on the update. This server provides additional comparisons between HMM profiles and the aforementioned CORES database.
LITERATURE server; allows users to search bibliography of interest in the topic.
An additional new tool in GyDB 2.0 is its wiki, powered by the MediaWiki content management system (http://www.mediawiki.org/). This tool has been implemented to allow other users participate in the project by editing or creating topics. Accession to this wiki is free but it requires a subscription (registration). The rationale behind this choice is that edits are registered by date and author in order to credit contributions, and secondly, we have programmed a revision mechanism to review all changes constructively before making them public. The top menu includes three sections to log in and manage the distinct wiki resources. Finally, to the right of the top menu, GyDB 2.0 includes a text field to search the whole project under two modes (detailed in Figure 1). The side menu divides the distinct GyDB sections into three major demarcations (emphasized with boxes in Figure 1). The first collects sections associated with the systematics applied at GyDB. The second implements information concerning the domains typically observed in the genomic structure of the elements we classify. The third demarcation offers free access to distinct databases, which are organized into three sections:
Trees and Networks; consists of the collection of inferred phylogenetic trees based on distinct protein domains encoded by the classified elements, or based on their concatenation (when they are parts of polyproteins). Remarkably, inferred pol polyprotein phylogenies based on the concatenation of the protease, reverse transcriptase, RNaseH and integrase domains, are the major criterion for assigning phylogenetic levels at GyDB 2.0 [results introduced in (7)]. Phylogenetic trees provide links to the corresponding element page at GyDB 2.0. By clicking any element name in any tree an entry assigned to this element is opened. These tree image maps were created using Phylograph 1.0 (15). This section includes the clan AA reference database (CAARD) of ancestral maximum likelihood (ML) reconstructions (13) that has been implemented and maintained at GyDB.
GyDB collection (16) or the repository of multiple alignments, HMMs, and majority rule consensus (MRC) sequences offered at GyDB 2.0. When a deposited alignment, profile or MRC sequence is associated with a journal publication, its entry in the collection includes citation information.
REF SEQ DATABASES or the repository for downloading the databases (GENOMES, CORES and LTRs) implemented in the BLAST server.
Finally, a variety of links to other database initiatives relevant to the topic are included in the side menu.
Figure 1.
GyDB 2.0 organization and implementation.
FUTURE PERSPECTIVES
Sequencing projects constantly deliver new types of MGEs [for example (17–22)]; hence the classification of non-redundant elements based on their phylogenetic signal is an open issue at GyDB, and results in the preparation of new sections. For example, we are committed to improving the understanding of the diversity and evolutionary dynamics of MGEs in eukaryotic and prokaryotic organisms. In this regard of eukaryotic LTR retroelements (the current database scope), the sequence repertoire at GyDB with representative elements retrieved from recently sequenced marine secondary endosymbionts including the brown alga Ectocarpus siliculosus (heterokont) and the coccolithophore Emiliania huxleyi (haptophyte) will be implemented. In terms of other research topics in preparation, one concerns the construction of a server devoted to the study of the complete set of MGEs and repeats (the mobilome) of biological genomes. This server will be introduced with two forthcoming publications focusing on the LTR retroelements and their related transposases of the pea aphid Acyrthosiphon pisum genome [see (23)]. At the technical level, we are exploring the application of formal grammars and machine learning algorithms to automate, as far as possible, the management and classification of the sequence data. We are also committed to developing solutions for other non-trivial difficulties that arise with the growing size of the databases. Viruses and MGEs usually show different rates of evolution and high variability depending on the evaluated protein or region. Therefore, we aim to implement more than one method of phylogenetic reconstruction to offer the user different perspectives based on different methods (or the opportunity to upload updated phylogenies via the wiki). On the other hand, the traditional view of the origin and evolution of biological systems is that they are usually monophyletic, but such an assumption has been challenged by increasing evidence suggesting that natural evolution can frequently proceed by gradual and vertical means, in addition to distinct modular, saltatory and reticulate events (24–36). In this respect, we are investigating appropriate protocols to combine phylogenetic inference with new tendencies in network biology [see also (7)].
FUNDING
Centro de Desarrollo Tecnológico Industrial (CDTI) (grant IDI-20100007, partial); Empresa Nacional de Innovación, S.A (ENISA) (17092008, partial); IMPIVA (IMIDTA/2009/118 and IMDTA/2010/740, partial); European Regional Development Fund (ERDF); Ministerio de Ciencia e Innovación (MICINN) (Torres-Quevedo grants PTQ-09-01-00020, PTQ-09-01-00670 and PTQ-10-03552, partial). Funding for open access charge: University of Valencia.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all the colleagues detailed in the list available at (http://gydb.org/index.php/Acknowledgments) for their support in contributing images of biological host organisms. We are also grateful to Senior NAR Editor Dr Michael Galperin and to the two anonymous reviewers for their constructive comments in improving this article. Finally we also thank Denys Wheatley and Angela Panther from Biomedes for copyediting of this article.
REFERENCES
- 1.Hurst GDD, Schilthuizen M. Selfish genetic elements and speciation. Heredity. 1998;80:2–8. [Google Scholar]
- 2.Volff JN, Brosius J. Modern genomes with retro-look: retrotransposed elements, retroposition and the origin of new genes. Genome Dyn. 2007;3:175–190. doi: 10.1159/000107611. [DOI] [PubMed] [Google Scholar]
- 3.Fauquet CM, Mayo MA, Desselberger U, Ball LA. Virus Taxonomy, VIIIth Report of the ICTV. London: Elsevier/Academic Press; 2005. [Google Scholar]
- 4.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 5.Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 2004;32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llorens C, Futami R, Bezemer D, Moya A. The Gypsy Database (GyDB) of mobile genetic elements. Nucleic Acids Res. 2008;36:38–46. doi: 10.1093/nar/gkm697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llorens C, Munoz-Pomer A, Bernad L, Botella H, Moya A. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol. Direct. 2009;4:41. doi: 10.1186/1745-6150-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorens C, Fares MA, Moya A. Relationships of Gag–pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol. Biol. 2008;8:276. doi: 10.1186/1471-2148-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 11.Koonin EV, Zhou S, Lucchesi JC. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llorens C, Futami R, Renaud G, Moya A. Bioinformatic flowchart and database to investigate the origins and diversity of Clan AA peptidases. Biol. Direct. 2009;4:3. doi: 10.1186/1745-6150-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llorens C, Futami R, Vicente-Ripolles M, Moya A. Biotechvana Bioinformatics. 2008. Phylograph: a multifunction Java editor for handling phylogenetic trees. Biotechvana, Valencia, SOFT: Phylograph. [Google Scholar]
- 16.Llorens C, Muñoz-Pomer A, Futami R, Moya A. Biotechvana Bioinformatics. 2009. The GyDB Collection of Viral and Mobile Genetic Element Models. Biotechvana, Valencia, CR: GyDB Collection. [Google Scholar]
- 17.Piskurek O, Nishihara H, Okada N. The evolution of two partner LINE/SINE families and a full-length chromodomain-containing Ty3/Gypsy LTR element in the first reptilian genome of Anolis carolinensis. Gene. 2008;441:111–118. doi: 10.1016/j.gene.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Novikova O, Mayorov V, Smyshlyaev G, Fursov M, Adkison L, Pisarenko O, Blinov A. Novel clades of chromodomain-containing Gypsy LTR retrotransposons from mosses (Bryophyta) Plant J. 2008;56:562–574. doi: 10.1111/j.1365-313X.2008.03621.x. [DOI] [PubMed] [Google Scholar]
- 19.Bae YA, Ahn JS, Kim SH, Rhyu MG, Kong Y, Cho SY. PwRn1, a novel Ty3/gypsy-like retrotransposon of Paragonimus westermani: molecular characters and its differentially preserved mobile potential according to host chromosomal polyploidy. BMC. Genomics. 2008;9:482. doi: 10.1186/1471-2164-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao D, Gill N, Kim HR, Walling JG, Zhang W, Fan C, Yu Y, Ma J, SanMiguel P, Jiang N, et al. A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J. 2009;60:820–831. doi: 10.1111/j.1365-313X.2009.04005.x. [DOI] [PubMed] [Google Scholar]
- 21.Gottlieb AM, Poggio L. Genomic screening in dioecious “yerba mate” tree (Ilex paraguariensis A. St. Hill., Aquifoliaceae) through representational difference analysis. Genetica. 2010;138:567–578. doi: 10.1007/s10709-010-9449-9. [DOI] [PubMed] [Google Scholar]
- 22.Maumus F, Allen AE, Mhiri C, Hu H, Jabbari K, Vardi A, Grandbastien MA, Bowler C. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics. 2009;10:624. doi: 10.1186/1471-2164-10-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik HS, Eickbush TH. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 1999;73:5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lerat E, Brunet F, Bazin C, Capy P. Is the evolution of transposable elements modular? Genetica. 1999;107:15–25. [PubMed] [Google Scholar]
- 26.Goodwin TJ, Poulter RT. A group of deuterostome Ty3/ gypsy-like retrotransposons with Ty1/ copia-like pol-domain orders. Mol. Genet. Genomics. 2002;267:481–491. doi: 10.1007/s00438-002-0679-0. [DOI] [PubMed] [Google Scholar]
- 27.Eickbush TH, Malik HS. Origin and evolution of retrotransposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 1111–1144. [Google Scholar]
- 28.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 29.Marco A, Marin I. How Athila retrotransposons survive in the Arabidopsis genome. BMC. Genomics. 2008;9:219. doi: 10.1186/1471-2164-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat. Rev. Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 31.Flavell AJ. Long terminal repeat retrotransposons jump between species. Proc. Natl Acad. Sci. USA. 1999;96:12211–12212. doi: 10.1073/pnas.96.22.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan IK, Matyunina LV, McDonald JF. Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc. Natl Acad. Sci. USA. 1999;96:12621–12625. doi: 10.1073/pnas.96.22.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bousalem M, Douzery EJ, Seal SE. Taxonomy, molecular phylogeny and evolution of plant reverse transcribing viruses (family Caulimoviridae) inferred from full-length genome and reverse transcriptase sequences. Arch. Virol. 2008;153:1085–1102. doi: 10.1007/s00705-008-0095-9. [DOI] [PubMed] [Google Scholar]
- 34.Koonin EV, Mushegian AR, Ryabov EV, Dolja VV. Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J. Gen. Virol. 1991;72(Pt 12):2895–2903. doi: 10.1099/0022-1317-72-12-2895. [DOI] [PubMed] [Google Scholar]
- 35.Llorens JV, Clark JB, Martinez-Garay I, Soriano S, deFrutos R, Martinez-Sebastian MJ. Gypsy endogenous retrovirus maintains potential infectivity in several species of Drosophilids. BMC Evol. Biol. 2008;8:302. doi: 10.1186/1471-2148-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Setta N, Van Sluys MA, Capy P, Carareto CM. Multiple invasions of Gypsy and Micropia retroelements in genus Zaprionus and melanogaster subgroup of the genus Drosophila. BMC Evol. Biol. 2009;9:279. doi: 10.1186/1471-2148-9-279. [DOI] [PMC free article] [PubMed] [Google Scholar]