Abstract
UK PubMed Central (UKPMC) is a full-text article database that extends the functionality of the original PubMed Central (PMC) repository. The UKPMC project was launched as the first ‘mirror’ site to PMC, which in analogy to the International Nucleotide Sequence Database Collaboration, aims to provide international preservation of the open and free-access biomedical literature. UKPMC (http://ukpmc.ac.uk) has undergone considerable development since its inception in 2007 and now includes both a UKPMC and PubMed search, as well as access to other records such as Agricola, Patents and recent biomedical theses. UKPMC also differs from PubMed/PMC in that the full text and abstract information can be searched in an integrated manner from one input box. Furthermore, UKPMC contains ‘Cited By’ information as an alternative way to navigate the literature and has incorporated text-mining approaches to semantically enrich content and integrate it with related database resources. Finally, UKPMC also offers added-value services (UKPMC+) that enable grantees to deposit manuscripts, link papers to grants, publish online portfolios and view citation information on their papers. Here we describe UKPMC and clarify the relationship between PMC and UKPMC, providing historical context and future directions, 10 years on from when PMC was first launched.
BACKGROUND
UK PubMed Central (UKPMC) is a free digital repository of biomedical and life sciences journal literature (http://ukpmc.ac.uk). It is based on PubMed Central (PMC), developed at the NCBI in the USA (1) and is part of a network of PMC International (PMCI) repositories that now also includes PMC Canada.
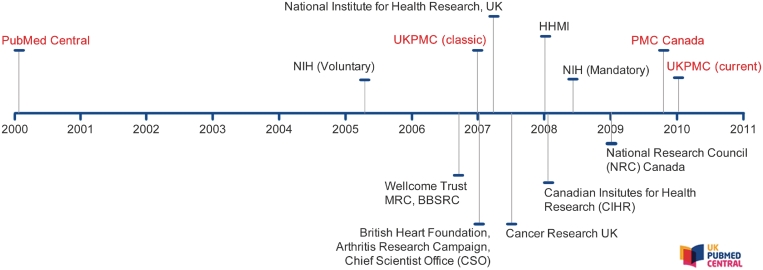
In 2006, the Wellcome Trust announced that research papers that had been funded by them must be made freely available via UKPMC no later than 6 months after publication (2). Working with other major funders of UK biomedical research, UKPMC was launched in January 2007 as a mirror of PMC (Figure 1). The funding agencies that support UKPMC are: Arthritis Research UK, BBSRC, British Heart Foundation, Cancer Research UK, Chief Scientist Office (Scotland), the MRC, National Institute for Health Research and the Wellcome Trust. Each of these funding bodies have a public access policy that states how publications arising from the research they fund have to be made publicly available in UKPMC. A list of websites listing each organizations’ public access policy can be seen here: http://ukpmc.ac.uk/Funders.
Figure 1.
Timeline of PMC USA, UKPMC and PMC Canada availability and related funding agency public access policies.
The mission of UKPMC is to become the information resource of choice for the UK’s life science and health research communities. To this end, it supports UK researchers in article deposition and grant reporting and provides access to the articles based on publicly funded research through a comprehensive electronic archive of the peer-reviewed literature relevant to the life sciences. The partner organizations charged with these tasks for UKPMC are the University of Manchester [Mimas and the National Centre for Text Mining (NaCTeM)], the European Bioinformatics Institute (EBI) and the British Library.
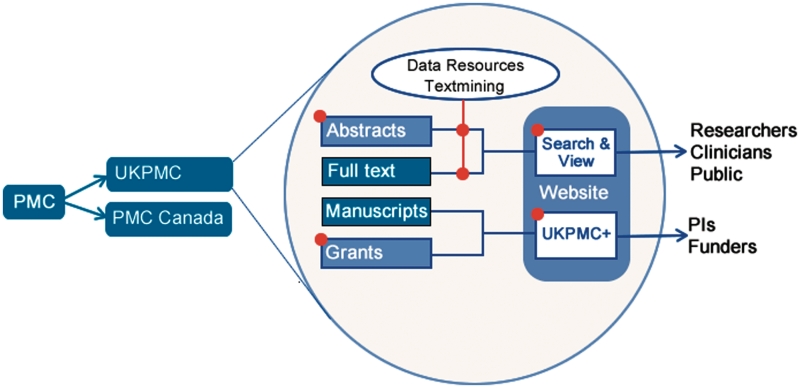
Since inception, UKPMC has developed tools for both researchers and funders that meet the specific requirements of the UK research community; these will be described later in this article. In terms of the archive itself, UKPMC is a ‘mirror’ of PMC USA, hosting the same content (with a few exceptions, see below), transferred daily from PMC in the USA to the PMCI nodes in the UK and Canada. Until recently, the article search and browse mechanisms used at all three sites were identical, supported by the software distributed as a component of the PMCI package. However, in January 2010, UKPMC launched a new interface that introduces novel features for navigating and searching the content. These include, for example, the ability to search biomedical abstracts and full-text articles from the same search box, the provision of different subsets of records such as patents and theses and the incorporation of citation data and text-mining-based applications. The new website and the related developments specific to UKPMC are the focus of this article; however we will set these developments within the broader context of PMCI (Figure 2).
Figure 2.
Overview of PMCI and specifically UKPMC services. The red nodes indicate areas in which UKPMC has extended the core PMCI installation, aside from the contribution of content.
THE RELATIONSHIP BETWEEN UKPMC AND PMC
PMCI is a collaborative effort between the NCBI (National Library of Medicine, National Institutes of Health, USA), PMCI nodes (UKPMC and PMC Canada) and the publishers whose journal content is archived in PMC. The vision of PMCI is to create a network of digital archives that share content in a manner analogous to the International Nucleotide Sequence Database Collaboration (GenBank/EMBL/DDBJ) model for data archiving and exchange across the globe (3–5). In this model, repositories in the network accept deposition of content locally, which is then distributed to the other repositories on a daily basis, as appropriate. This has the advantage of maintaining data integrity and the viability of the archive through replication across several different global sites. Furthermore, the ability for all sites to ingest content engages the local user community and allows location-specific requirements to be addressed. This model also encourages innovation: using the same base set of data, novel access methods or uses of the data can be developed.
To date, all the content in UKPMC is routed via PMC, even manuscripts submitted locally via the UKPMC Manuscript Submission System (see below). Once added to PMC, new content is pushed to UKPMC daily, from where articles are displayed using the PMCI software supplied as a bundle with the PMC database. Using the same rendering engine to display articles ensures that article content is faithfully displayed at all sites, having undergone rigorous quality assurance and sign-off by a combination of PMC in-house staff, authors and/or the publishers that supplied the article. Therefore if you compare the same article displayed in both PMC and UKPMC, the websites look different and offer different functionality, but the core of the article is exactly the same.
CONTENT GROWTH
When PubMed Central was first launched in 2000 (1), it was with a nucleus of only a few participating journals such as the ‘Proceedings of the National Academy of Sciences USA’ and ‘Molecular Biology of the Cell’. Over the past 10 years, these few pioneers have grown to a list of over 2000 titles whose content is represented in the archive. At the time of writing, there are over two million articles available in PMC, about 1.8 million of which are distributed to UKPMC and PMC Canada.
How does content get into UKPMC?
With the growing content base there has been an increased variety of ways in which content gets deposited. These are described below:
- The journal makes 100% of its content available in the archive
- at the time of publication; these are usually Open Access journals, but not always.
- with a specified time delay after the publication date. The time delay is stipulated by the publisher and does not necessarily coincide with the time delay specified by funder public access policies. These are usually Free Access articles.
- The journal deposits content on an article-by-article basis
- because an author has chosen an open access track publishing option, (for example as a result of a funder’s requirement for public access); Open Access tracks are now offered by a number of journals.
- the journal has agreed to deposit—for no fee—the final published article on behalf of an author funded by one of the UKPMC funders, NIH or HHMI. In such cases, articles are typically embargoed and are not included in the Open Access subset.
-
(3)
The final, accepted manuscript (not the published PDF) of an article is deposited via the Manuscript Submission System by the author (‘self-archiving’). This occurs when a researcher has to deposit an article as a condition of being funded by an agency with a public access policy and the journal agrees to that requirement, but the journal does not have an in-house publication option to deposit the article on the author’s behalf.
-
(4)
The article is available as a part of the PMC Back Issue Digitization Project. In this project, journals that joined PMC prior to 2008 had the opportunity to have back-copy paper-based content scanned. This was a joint project between NLM, the Wellcome Trust and the UK Joint Information Systems Committee (JISC) and has resulted in ∼1.2 million articles in the archive, although some of these are not available in UKPMC.
Are all the articles in UKPMC open access?
All of the articles in UKPMC are ‘Free Access’ which means that they are free for anyone to search, view, read and download in PDF form, if available. However these articles are still protected by publisher copyright and cannot be reused in any way for research (e.g. text mining) or commercial purposes without the explicit permission of the copyright holder. For these reasons, UKPMC content cannot be redistributed and the display of articles is tightly regulated.
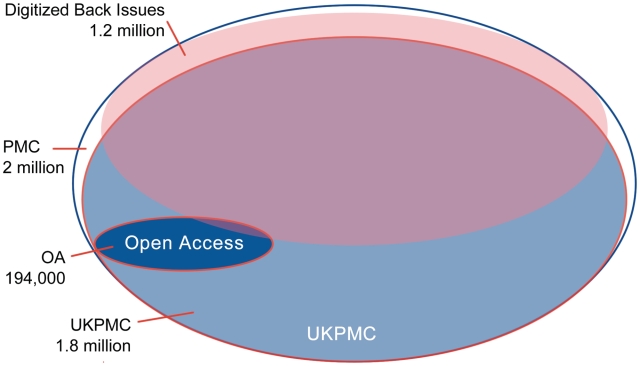
About 10% (over 190 000) of all the articles are ‘Open Access’, which means that they can be used in any way (e.g. for text mining) as long as the original authors and journal source are acknowledged (Figure 3). (Although, some open access licenses contain some restriction, for example to non-commercial use). These articles, complete with associated graphics files, are available for FTP download from here: http://www.ncbi.nlm.nih.gov/pmc/about/ftp.html
Figure 3.
Content profile of PMC and UKPMC. This demonstrates that ∼90% of PMC content is made available to UKPMC. Roughly 60% of all articles are from digitized back issues; these represent the bulk (∼80%) of the content not available in UKPMC. The proportion of content available as XML is increasing as the database grows, being the standard format for active deposition of new articles. The Open Access articles make up almost 10% of the total content; over 90% of these are available as XML.
An XML files-only download of Open Access articles is also available from here: http://ukpmc.ac.uk/ftp/oa
Differences between PMC and UKPMC content
As described earlier, not all content available in PMC is made available to UKPMC and PMC Canada (Figure 3). Since July 2006, all PMC Participation Agreements have included permission to make a participating journal’s PMC content available at UKPMC and since June 2009, to PMC Canada also. However, while most publishers participating in PMC prior to July 2006 agreed to deposit their content in UKPMC, some did not. This means that some 250 000 articles are not part of the PMCI Agreement, the vast majority of which, about 190 000, are articles from the Back Issue Digitization Project. Content not currently available in UKPMC is listed here: http://ukpmc.ac.uk/ppmc-localhtml/not_in_ukpmc.html
THE UKPMC WEBSITE AND SERVICES
The partner organizations responsible for UKPMC hosting and development are the University of Manchester (Mimas and NaCTeM), the EBI and the British Library. In close collaboration with the NCBI, these partners have developed the website, the search and retrieval system, integrated text-mining-based features, manuscript submission and grant reporting tools. All these tools and features are available from: http://ukpmc.ac.uk.
The UKPMC archive is supplemented by the CiteXplore citations database (http://www.ebi.ac.uk/citexplore) and both can be searched directly from the homepage. Further links to UKPMC+ (manuscript submission and grant reporting), FAQs and information about the project are also provided.
User-centered development
To inform the development of services, UKPMC conducted a user survey in 2008 to review user attitudes and requirements. The feedback highlighted several key areas where enhancements to the service were desirable:
Improving information retrieval and knowledge discovery through the development of text and data-mining solutions;
Identifying and providing access to additional non-journal content; and
Creating tools to enable users to track research grants and the research papers associated with them.
The results of the survey and a follow-up workshop have translated into some of the new developments now available at the UKPMC website.
UKPMC has undertaken a public engagement campaign to present emerging developments to focus groups of life science researchers, promote the use of the service within UK Higher Education Institutions and encourage article deposition by researchers. Along with directives and feedback from the UKPMC Advisory Board, these activities have been critical to ensure the continued relevance of UKPMC to its core constituency.
Additional content available at the UKPMC website
The UKPMC website enables single-point access for the search and retrieval of both the full text content of UKPMC and the content of CiteXplore, the citations database developed at the EBI. CiteXplore contains metadata only and includes all of PubMed's 20 million and more abstracts, around 0.5 million records from Agricola and 3.4 million records from the European Patent Office. Further content has also been added to CiteXplore in response to user feedback, which requested that other relevant non-journal content be made available. This additional content includes over 40 000 theses and 2700 clinical guideline documents that have full-text links, sourced from the UK National Health Service (NHS). Furthermore, CiteXplore also contains the metadata for about 400 000 UKPMC full text records that are not represented in PubMed.
Using UKPMC
Core search and retrieval
When a search term is typed into the search box on the homepage, both the UKPMC full text archive and the CiteXplore citations database are automatically searched. The default view shows the results found from the citations search (indicated by ‘All Citations’ and the number of results in red) in publication date order. The results from the full text UKPMC search can be viewed by clicking on the ‘Full text articles’ link, which also displays the count of results. Popular content subsets are highlighted on the right hand side of the page; these include, for the citations search: reviews, clinical trials, systematic reviews, patents, theses and the NHS clinical guideline documents mentioned previously. Any citation for which there is full text in UKPMC is highlighted by the UKPMC logo.
All results lists can be toggled for browsing purposes; furthermore, the default sort order of publication date can be changed to ‘relevance’ sort, which is based on the frequency of the term appearing in the document, compared with the corpus as a whole. This relevancy ranking is a core function of Lucene, the open source software on which the search index is based. We have further extended the basic Lucene query handling and logic to incorporate:
use of the usual terminology for Boolean (AND, NOT, OR), phrase (``'') and wildcard (*) searching;
stemming of search terms in the title, abstract and keyword fields only;
a query expansion function that expands your search terms to its known synonyms in MeSH and UniProt gene symbols (switched on by default in UKPMC searches); and
special handling of author name searches that gives additional weight to terms found in the author field, promoting them to the top of the search results
The abstract for any citation or full text article can be viewed either by clicking ‘More’ at the end of each result or by clicking on the title of the article. Clicking on an author name or journal title will elicit searches for or refine the current search on those entities.
In a given session of searching and browsing, previous searches and article views can be revisited using the Recent Activity function, found on the top right of every search page. Clicking on this icon pops up a list of all searches and pages viewed in the past 24 h, providing a useful overview of the navigation history across search sessions.
The Clipboard, located next to the Recent Activity, used in conjunction with the red Clipboard icons next to each result returned, allows the user to collect citations and export them in the Research Information Systems (RIS) format widely used by reference managers including EndNote, Mendeley and ProCite. In addition to citations, the Clipboard can also be used to collect search queries, ‘Cited By’ citations, text-mined biological terms (Bioentities) and Related Articles (see below). Snapshots of the Clipboard can be exported as HTML either to file or email. These exported Clipboard snapshots contain a link that enables the user to resume their previous Clipboard session at a later date, even from a different computer.
Viewing abstracts and full text articles
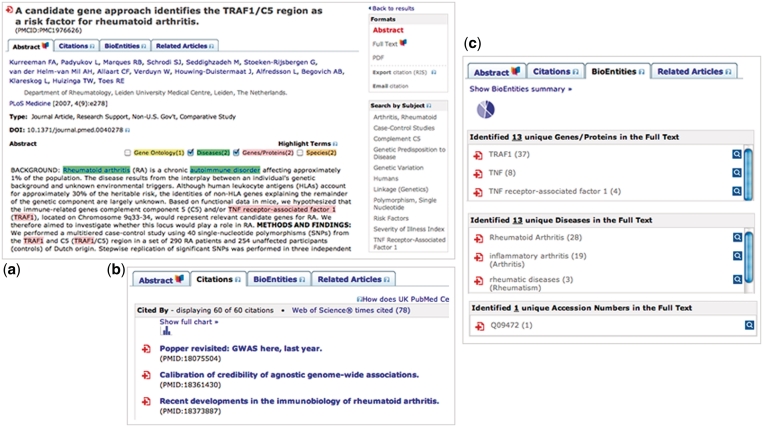
Having clicked on an article title from the search results, the complete citation record is viewed (Figure 4). Each citation is displayed with:
links to the full-text article in UKPMC or on the publisher’s website, where available;
a Citations Tab, containing information on the references listed in the article and a list of articles citing the current one, along with counts, where available;
a Bioentities Tab, containing a summary of terms text-mined from the full text article, linked to as well as links to databases such as UniProt, PDB and Entrez Gene;
Related Articles Tab, populated via NCBI eUtilties. (These are the same related articles as seen in PubMed.);
a ‘Highlight Terms’ function on the abstract. This function uses the same back-end processes as used to generate the text-mined terms list in the Bioentities Tab, with links to similar databases. The terms are colour-coded by entity type (listed below); and
a list of subject terms based on MeSH terms (when available) that can be used to initiate new searches or refine your current search. In the case of a full-text article view, this MeSH-based list is replaced with an abbreviated form of the text-mined terms list displayed in the Bioentities Tab.
Figure 4.
Some key features of the UKPMC website. (a) The abstract display has links to the full text, tabs that contain Citation, Bioentities and Related Articles information and a ‘highlight terms’ function on the abstract. (b) View of the citations tab, showing citing articles. (c) Extract from the Bioentities tab, showing mined gene/protein names, diseases and Accession numbers.
All these features, in concert with the Clipboard and Recent Activity facilities, offer alternative ways to browse and search the content, providing integrated scientific context as well as complementary means to assess the relevance of an article for your needs.
The application of text mining in UKPMC
The text-mined terms lists located in the Bioentities Tab and the term highlighting function in abstracts are based on the application of text mining to the CiteXplore and UKPMC collections, thanks to expertise supplied by the collaborative efforts of the EBI and NaCTeM.
Several broad categories of terms have been extracted by means of Named Entity Recognition algorithms that leverage a variety of vocabularies to identify those terms in full text articles. At the time of writing, these are:
genes/proteins;
organisms;
Gene Ontology (GO) terms;
diseases;
Accession numbers; and
chemicals.
The mined terms are made available as summary tables, as described above, with the genes/proteins, organisms and GO terms linked to UniProt; Accession numbers linked to the EMBL Nucleotide Archive, UniProt or PDB; chemicals linked to ChEMBL; and disease terms reissuing a literature search. The text-mined term summary tables also shows a count of the frequency of occurrence of the category as a whole as well as a count of how often individual terms appear in the article. These same terms are also indexed, allowing a user to limit keyword searches to the specific mined-term fields. As of July 2010, there are over 40 million text-mined annotations in UKPMC full-text articles, covering over half a million unique terms.
Citation information
The Citations Tab contains two article lists: ‘Cited By’, articles that have cited the current article and ‘Cites the Following’, articles in the reference list of the current article. The construction of the ‘Cited By’ list requires access to article reference lists, plus the ability to resolve those references to an unequivocal source (for example a PubMed ID). In this way, reference lists from articles published subsequent to the current one can be processed to provide a list of ‘Cited By’ articles and thereby one indication of the impact of the article.
The number of ‘Cited By’ articles listed is to some extent a factor of the total number of articles in the data set used to calculate it. The UKPMC citation network is calculated from the UKPMC content, supplemented with metadata supplied by CrossRef (http://www.crossref.org). While it is the largest citation network available in the public domain, the amount of content available to UKPMC is less than that used in commercially available services. The citation counts displayed on the UKPMC website are therefore often lower than those from commercial products; for comparative purposes, counts from Thompson-Reuters' Web of Science are listed alongside the UKPMC counts.
UKPMC+: tools for UK funded researchers
While most of the UKPMC website is open for anyone to use, some features are restricted to the funding agencies that support UKPMC and the Principal Investigator (PI) on grants awarded by those funders. These services relate to manuscript submission and grant reporting (Figure 5). The exception to this is the ‘Grant Lookup’ facility, available on the main UKPMC website. Here, anyone can search and retrieve information on grants awarded by the UKPMC Funders. However, the remainder of the tools described in this section are available only to researchers funded by supporting funding bodies via UKPMC+, at http://ukpmc.ac.uk/ukpmcplus.
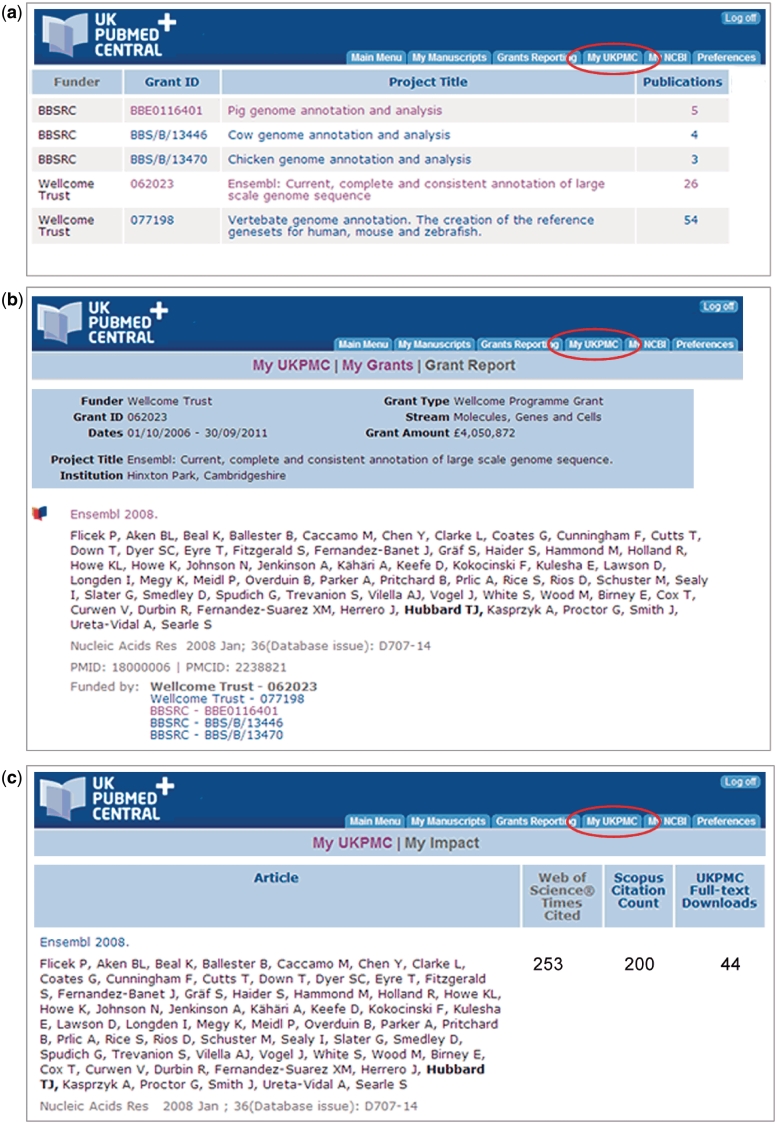
Figure 5.
Key features of the Grant Reporting system unique to UKPMC+. After grants have been linked with published papers via the ‘Grant Reporting’ pages, the My UKPMC tab offers detailed reports on these relationships, along with citation information on the listed papers. (a) Grants Summary list, showing the number of papers associated with each grant. (b) Detailed listing of publications associated with a particular grant. Note that the articles also display other grants associated with the article. (c) The My Impact report. This shows the number of times each article has been cited in Web of Science and Scopus. The number of downloads from the UKPMC website are also reported.
Manuscript submission service
UKPMC+ provides a manuscript submission service to support authors depositing articles in order to comply with the UKPMC Funders public access policies. It extends software distributed via PMCI, originally developed for the NIH Manuscript System and allows users to deposit and manage their final, peer-reviewed manuscripts, in line with journal policy. Manuscripts in a wide range of electronic formats can be submitted along with figures, tables or supplementary data.
A username and password is required to access the service. These login details are provided to PIs automatically, by Email, once their grant has been awarded. Other users, such as administrators, can use the system to create their own accounts and submit on behalf of PIs. Papers submitted through UKPMC+ are made accessible through all PMCI nodes. The UKPMC+ manuscript submission service also allows users to attach their grants to existing PubMed and PubMed Central papers via the ‘Grant Reporting’ facility and is integrated with MyNCBI.
The UKPMC+ submission service is supported by a dedicated Helpdesk Team who manage each submission, from deposit through to publication on UKPMC. The Helpdesk is also there to provide support on the full range of UKPMC services. (Contact the Helpdesk at ukpmc@bl.uk or on +44 [0]1937 546699.)
Grant reporting tools
Within UKPMC+, the existing MyNCBI function has been supplemented by My UKPMC, which offers reporting for individual authors, including unique reports that link the PIs portfolio of grants with the associated research articles and those that report the impact of those articles (Figure 5):
‘My Grants’ shows a PI’s known grants and the associated publications, which link to the full-text articles in UKPMC. A grant report can be ‘published’, i.e. made available at a persistent URL, suitable for inclusion in an end-of-grant report.
‘My Impact’ shows the impact of a PI’s research as a list of the reported published articles along with citation counts from Web of Science and Scopus and a count of article downloads from the UKPMC website. The citation counts are calculated daily and link through to the relevant pages in Web of Science and Scopus. This too can be ‘published’ as a static web page for inclusion on author’s personal web site, for example.
FUTURE DIRECTIONS
The past 5 years have seen UKPMC grow from a basic mirror of PMC into a complementary resource offering additional types of content and search features based on open source software and text mining techniques. This work lays the foundations for future content growth and the development of new tools to help life science researchers discover relevant information faster, within the context of other public data resources.
Research articles will continue to be the primary unit of currency for the archive; however, we plan to make other biomedical full-text resources available as part of the service. Building Open and Free Access content deposition in UKPMC will require continued support for researchers to reach the goal of 100% compliance with all UK Funder public access policies. To this end, we will continue to improve the grant reporting tools available to the UK research community.
Looking beyond the UK, extending UKPMC into a Europe-wide resource will provide further opportunities for growth; UKPMC already has four associate European Funders that support article deposition into UKPMC on a manuscript-by-manuscript basis for their grant holders. These are Telethon Italy, Science Foundation Ireland, Austrian Sciences Research Fund and the Health Research Board, Ireland.
The text-mined annotations currently available in UKPMC are the starting point for the development of innovative literature search and browse tools embedded within public domain data resources. For example work is ongoing to identify relationships between bioentities at the sentence level, in order to build tools that help the user focus more rapidly on areas of interest. The current programme of work demonstrates the quality and scope of the core text mining; where possible, we aim to share these data enrichments along with the article content with the scientific, library and publishing communities.
In the tradition of UKPMC's commitment to community engagement, we invite you to explore the UKPMC website and welcome feedback on all aspects of the project.
FUNDING
Funding for open access charge: UKPMC Funders (led by Wellcome Trust).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the UKPMC Board Members, 2006–2011: Michael Ashburner University of Cambridge (until 2008), Casey Bergman University of Manchester; Anne De Roeck Open University; Alison Henning Wellcome Trust; Colin Hopkins Imperial College (until 2008); Tim Hubbard Wellcome Trust Sanger Institute; David Ingram (Chair) University College London; Douglas Kell BBSRC (until 2008); Robert Kiley Wellcome Trust; Peter Murray-Rust University of Cambridge; Tony Peatfield Medical Research Council; Philippa Saunders MRC Human Reproductive Sciences Unit; Lynne Roberts Warwick University (until 2008); Frank Uhlmann Cancer Research UK.
REFERENCES
- 1.Roberts RJ. PubMed Central: The GenBank of the published literature. Proc. Natl Acad. Sci. USA. 2001;98:381–382. doi: 10.1073/pnas.98.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walport M, Kiley R. Open access, UK PubMed Central and the Wellcome Trust. J. Roy. Soc. Med. 2006;99:438–439. doi: 10.1258/jrsm.99.9.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaminuma E, Mashima J, Kodama Y, Gojobori T, Ogasawara O, Okubo K, Takagi T, Nakamura Y. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 2010;38(Database issue):D33–D38. doi: 10.1093/nar/gkp847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leinonen R, Akhtar R, Birney E, Bonfield J, Bower L, Corbett M, Cheng Y, Demiralp F, Faruque N, Goodgame N, et al. Improvements to services at the European Nucleotide Archive. Nucleic Acids Res. 2010;38(Database issue):D39–D45. doi: 10.1093/nar/gkp998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nuclear Acids Res. 2010;38(Database issue):D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]