Abstract
The Ensembl project (http://www.ensembl.org) seeks to enable genomic science by providing high quality, integrated annotation on chordate and selected eukaryotic genomes within a consistent and accessible infrastructure. All supported species include comprehensive, evidence-based gene annotations and a selected set of genomes includes additional data focused on variation, comparative, evolutionary, functional and regulatory annotation. The most advanced resources are provided for key species including human, mouse, rat and zebrafish reflecting the popularity and importance of these species in biomedical research. As of Ensembl release 59 (August 2010), 56 species are supported of which 5 have been added in the past year. Since our previous report, we have substantially improved the presentation and integration of both data of disease relevance and the regulatory state of different cell types.
INTRODUCTION
Ensembl provides high-quality genome annotation across chordate species through a comprehensive set of methods, which result in unique data sets including Ensembl’s gene annotations, multiple alignments, gene homology relationships and regulatory annotation. We also integrate data resources available in species or domain specific databases such as ZFIN (1), HGNC (2), the Single Nucleotide Polymorphism Database (dbSNP) (3), UniProt (4) and the Encyclopedia of DNA Elements (ENCODE) portal at UCSC (5). We continue to work closely with major providers of genome information and participate in primary analysis of newly sequenced genomes including zebra finch (6) and turkey (7).
Most users access Ensembl though our genome browser available at http://www.ensembl.org or by downloading our data sets for use in specific analysis contexts. In addition, Ensembl data and source code are provided freely to all of our users and we encourage incorporation of our code base and data into third party resources as well as the programmatic use of our results in external analysis pipelines that interact with these results using the Ensembl API (8), Ensembl tools such as the SNP Effect Predictor (9) and the Ensembl BioMart (10).
This article provides an overview of some of the new data and features that have been added to Ensembl since our previous report (11) and provides details of changes to our integrated analysis procedures that are designed to maximize the value of new and emerging technologies such as RNA-seq and ChIP-seq. Detailed documentation of Ensembl’s data and software is available from our web site. We also published a series of articles in the past year describing many of Ensembl’s methods in detail (12–17), and these will be referenced as appropriate below.
RESULTS
Ensembl is updated several times each year with new species and updated genome assemblies. In addition to new data sets, updates normally include software and visualization enhancements which are designed to both improve our existing codebase and provide support for the new data types. We have seen a continual growth in the size and complexity of each release over and above the contribution of newly supported species. We have developed a number of techniques to adequately manage this complexity and ensure that our users remain able to access our data efficiently. These developments aim to ensure that new data types can be seamlessly integrated into Ensembl.
Variation
Ensembl variation has been extended this year to provide much more information regarding disease and phenotypic annotations on sequence polymorphisms and in the context of somatic mutations. For example, we have nearly 60 000 phenotype annotations from our growing resource of Genome Wide Association Study (GWAS) data. We have also incorporated real time information into our variation-based web displays from SNPedia (http://www.snpedia.com/index.php/SNPedia) using the Distributed Annotation System (DAS) protocol (18). For germline mutations, we include the location and identifier of mutations from the Human Gene Mutation Database (HGMD) (19) as well as sequence somatic mutations from genes in the Catalogue of Somatic Mutations in Cancer (COSMIC) (20).
Addressing the data created in clinical and diagnostic contexts, as well as data available in Locus Specific Databases (LSDBs), we have worked with several partners to develop standard, common reference sequences, called ‘Locus Reference Genomic’ sequences (LRGs) (21). LRGs are designed to ensure stability of data reporting while facilitating the integration of variation and mutation data into genome-wide resources. The lack of reporting stability in the context of changes to the genome assembly has previously impeded data exchange. We have significantly adapted existing Ensembl core API functionality to facilitate the storage of the structure and sequence of LRGs in parallel with the reference sequence and annotation features. The LRG displays within Ensembl are able to access this specific sequence data and display variation data submitted using the LRG coordinate systems. Taken together, the LRG infrastructure in Ensembl brings together specific diagnostic and LSDB data in such a way as it can be integrated and viewed alongside the current resources. The data will have searchable links to the originating LSDB providing mutual benefit to the resources.
Ensembl variation databases continue to be updated with short sequence polymorphisms from dbSNP whenever a new release is available. Recent dbSNP releases have contained extensive early results from the 1000 Genomes Project that will eventually provide a reference set of all common human variation in several different populations down to 1% minor allele frequency. Structural variation, including copy number variants, is imported from the collaborative Database of Genomic Structural Variations (dbVar) and Database of Genomic Variants archive (DGVa) projects (22) for human, mouse, pig and dog. A full description of our process for creating the Ensembl variation databases as well as a detailed description of each of the Ensembl variation displays was recently published (16). A companion publication described the variation database and software infrastructure (13).
On the website we have added a new display, ‘Linked Variation’ to the variation pages to show a list of variants and their associated phenotypes that are in high LD with the display variant (Figure 1). We have also expanded the context panel to show structural variants, regulatory regions or highly conserved elements that overlap the variant. If the sequence displays are configured by the user to show variation data, there are now enhanced pop-up windows which appear by clicking on a variant to display population frequency data and also provide direct links to overlapping genes and transcripts. It is also possible to filter the sequence displays to hide variants in a specific population below a particular minor allele frequency from the ‘configure this page’ link. For phenotypic data, we have a new display highlighting variants associated with a particular phenotype across the whole karyotype and colour coded these variants by P-value.
Figure 1.
A view of variants with high linkage disequilibrium to rs1333049 in the Tosconi (TSI) population including the phenotypes associated with these variants and colour-coded P-values representing the strength of these phenotype associations. Variants have been sorted by distance to the focus SNP using the arrows in the table header row.
To help users interpret their own data, Ensembl now provides the SNP Effect Predictor both on the website and as a downloadable Perl script (9). This tool accepts a simple tab-delimited data file of SNP and indel changes, from the user and has proved very popular. It outputs the predicted consequence of these variants, based on the annotation contained in Ensembl. This includes whether they fall in a transcript, the amino acid position and change (if the variant falls within a protein) and the variant identifier of known variants that occur at this position.
Regulation
Ensembl’s regulatory information has seen substantial increases in the quantity and variety of data stored over the past year. Widespread uptake of high-throughput sequence based methods for assay of chromatin-based samples in projects including ENCODE (23) and the Epigenomics Roadmap (http://nihroadmap.nih.gov/epigenomics/), as well as smaller hypothesis-driven projects, has initiated a flood of data on transcription factor binding and chromatin state. As of Ensembl release 59 (August 2010) the functional genomics database has incorporated 285 data sets from these projects primarily from ChIP-seq and DNase-seq assays for human and mouse cells. Data on the genomic location of binding sites for 56 transcription factors (TFs) as well as the locations of sites for 40 modified histones has been incorporated. An additional 23 data sets that identify sites of open chromatin or DNase I hypersensitivity are also now available. These data cover 9 human and 4 murine cell types, and are incorporated via a standardized read-mapping and peak calling pipeline to generate both normalized signal data and predicted enriched regions (‘peaks’ or ‘hits’) with peak summits. Signal data is stored in a compressed binary format. The pipeline also incorporates filtering steps to remove artifactual enrichments known to be generated during sequence alignment of these data. For TF data with verified position weight matrices in the JASPAR database of transcription factor binding profiles (24), the positions of significant [at 0.05 empirical False Discovery Rate (FDR)] putative binding sites within the hit regions are also presented. We will continue to incorporate new data in this area as it becomes publicly available.
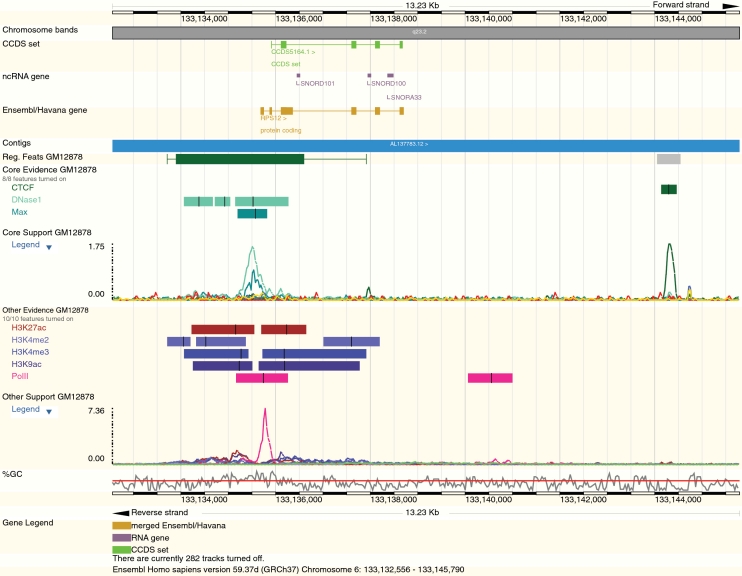
The past year has also seen major developments to the Ensembl Regulatory Build. In its revised form, the regulatory build process uses all TF and open chromatin data across all cell types to establish locations active in regulation in a multi-cell build step (referred to as ‘core regulatory features’). Each core region is then interrogated on an individual cell type basis to extend the region where supporting data is present in that cell type. Regulatory features are annotated by the nature of the data present within a given feature. This process gives a set of regulatory features for each cell type, as well as a set of core regulatory features presented in a multi-cell track. Cell-specific and multi-cell regulatory features can be viewed in both the location view and in the specialized regulation panels, together with the underlying signal and hit regions (Figure 2). To provide Regulatory Build annotation on cell types without TF and open chromatin data we have developed a conservative projection version of this build that projects regulatory features from the multi-cell build onto cell lines where less information is available.
Figure 2.
Core and other evidence regulatory features near the 5′-end of the RPS12 gene in the GM12878 cell line. A representation of the raw DNAse-seq and ChIP-seq signals are shown for both types of features in the multi-wiggle plot below the respective collection of significant hits. Peak centres are marked in each hit by a vertical bar.
To complement the new data and Regulatory Build there have been substantial improvements to our regulatory views. In particular, we have introduced a new type of track to allow display of multiple sets of signal data within the same track, the ‘multi-wiggle’ track (Figure 2). Multi-wiggles can be displayed in location view or as part of the Detail panel of the Regulation specific views. Currently data within multi-wiggle tracks is organized by cell type, with data split into core evidence (TFs and open chromatin used to identify regulatory feature cores) and supporting evidence (histone modifications). The same data types have the same colour in each track. Within the Regulation views there is considerable information to reflect the new cell specific nature of the data and Regulatory Build. In each case, control of the data to be displayed is via a revised panel accessible from the ‘configure this page link’.
The functional genomics database continues to provide mapping of probe sets for all the common microarray platforms, as well as a standalone environment to support probe set mapping. A detailed description of the annotation pipeline for probe sets and an analysis of the quality of the results were recently published (17).
Gene annotation
Over the past year, we have focused on continued improvements to the annotation of the human genome as well as the development and testing of a de novo RNA-seq gene annotation pipeline to a point where it is suitable for annotating novel genes in the zebrafish zv9 assembly.
Gene annotation on the human genome assembly GRCh37 is updated with each release to include the latest Havana annotations as part of GENCODE (25), which is a project of ENCODE (23). Since Ensembl release 56 (September 2009), the Ensembl human gene set has exactly corresponded to a GENCODE release. GENCODE releases also contain the full set of consensus protein coding translations identified by the consensus coding sequence (CCDS) project (26). The algorithm and code base used to create this merged, consensus gene set from the Ensembl and Havana gene sets have developed and matured over the past year, which has strengthened our collaboration with Havana and is leading to the production of the best possible gene set for human. For example, all gene types/biotypes annotated by Havana are now included in the Ensembl-Havana consensus set, with the Havana biotype taking priority at loci where both groups have produced annotation.
This year also saw the first release of new non-reference human assembly patches based on the reference GRCh37 assembly. These patches are produced by the Genome Reference Consortium (GRC: http://www.genomereference.org) and one of the patched regions included the ABO gene, which was known to have an impossible haplotype in the reference assembly. Ensembl release 59 (August 2010) incorporated the first set of patches with basic annotation on these new regions. These patches are stored within the Ensembl core database, and are applied on-the-fly as required. The same functionality that supports haplotypes and pseudoautosomal regions is used to support the assembly patches, including patches that add novel sequence and patches that modify existing sequence. The GRC recently released a second set of patches to the GRCh37 assembly and further patch releases are expected in the future. We plan to continue integrating the patches and providing basic annotation on them.
Further improvements to human genome annotation have come via our newly developed lincRNA annotation pipeline. This procedure predicts long intergenic non-coding transcript models using both cDNA alignments and ChIP-seq data. In addition to these developments, the human Expressed Sequence Tag (EST) alignments have been recently updated and the database holding these alignments has been optimized in order to increase the speed at which the main website displays these results.
The annotation of the mouse genome has benefited from the above developments for human, with mouse now also including lincRNAs. The consensus mouse gene set continues to be a merge of Ensembl and Havana annotation, incorporating improvements developed as part of the GENCODE merge process. We continue to work with the CCDS project for both human and mouse gene sets (26) using the gene models described here as our input to the project and identifying those genes in our databases that are part of the CCDS sets.
Our other major development effort over the past year has been the continued optimization of our new annotation pipeline that uses only RNA-seq data as input to create transcript models. The refined RNA-seq annotation pipeline was used in the annotation of the zebrafish zv9 assembly and earlier versions of this pipeline were used to annotate human, worm and fly data for the RNA-seq Genome Annotation Assessment Project (RGASP) 1.2. The zv8 assembly provided a platform for much of the development of the pipeline and the Ensembl website now displays a number of informative DAS tracks, including transcript models built from a range of tissues and also expression information in the form of intron alignments.
Beyond these new developments in our gene annotation and strategy, we have included a number of new species and updated genome assemblies into Ensembl over the past year. The Ensembl Pre! site saw the addition of five new species: baboon (Papio hamadryas), turkey (Meleagris gallopavo), duck (Anas platyrhynchos), panda (Ailuropoda melanoleuca) and sheep (Ovis aries). In addition, the new zebrafish zv9 assembly has been made available on the Pre! site and will soon be released on the main site. On the main Ensembl web site, we have included updated and reannotated assemblies for elephant (Loxodonta africana), rabbit (Oryctolagus cuniculus), marmoset (Callithrix jacchus) and gorilla (Gorilla gorilla). All low-coverage species annotated by the Ensembl projection-build pipeline have also been updated to improve the annotation of selenocysteine amino acids. We have also developed a new method for closing gaps in annotations (i.e. false introns) generated where an aligned protein does not match the genomic sequence.
Comparative genomics
As the number of supported genomes in Ensembl grows, so do the computational demands of our comparative genomics resources. We have made a significant effort to consolidate and automate our data production pipelines to enable them to continue to scale (14). These developments have allowed us to expand the set of data we provide and these expansions are described below.
The Ensembl GeneTrees provide homology relationships between genes annotated on Ensembl supported species (27). These have been extended in the past year in two ways. First, our gene trees now include short ncRNA genes that are generally much shorter than the protein-coding genes that the method was originally developed for. This required a modification of the original protein-coding centric pipeline to include the flanking region of the ncRNA genes and, therefore, increase the specificity of the alignments. Second, we have recently introduced the concept of ‘possible orthologs’, which are usually ill-supported between-species paralogs. These cases are typically found where a weakly supported duplication results in orthologs being wrongly called as between-species paralogs. We find this category especially useful and relevant in cases where the tree does not show any ortholog for one particular species. New features have also been added to the GeneTree viewer. For instance, new tick marks representing introns have been added to the alignment overview and it is now possible to highlight ortholog pairs on the tree.
The family of Ensembl whole-genome multiple alignments has been extended to include a new fish-specific set of multiple alignments and an expanded set of primate multiple alignments incorporating gorilla and marmoset. Aligning fish genomes is a complex task due to the larger evolutionary distance among them compared to placental mammalian genomes and required some adaptations to the EPO (Enredo-Pecan-Ortheus) pipeline (28–30) used for our other multiple alignments. As for the placental mammalian genomes, we run Genomic Evolutionary Rate Profiling (GERP) (31) on the fish genome alignments to produce both per base conservation scores and constrained elements.
Beyond the whole-genome multiple alignments, we now provide aligned mitochondrial genomes via a specific alignment pipeline as these were not included in the original EPO alignments. Support for the assembly patches described above that are now provided on the latest human assembly has also been incorporated as have a larger collection of pairwise alignments such as pig-cow and wallaby-opossum alignments.
Ensembl core software and data access
To improve website performance for users, we have created a second mirror of the Ensembl website in the USA. The new mirror site, on the east coast of the USA is located at http://useast.ensembl.org/ and joins our existing mirror site at http://uswest.ensembl.org/. The new mirror site is noteworthy in that it uses the Amazon Web Services (AWS) cloud computing infrastructure rather than dedicated hardware in a co-location facility. We intend to continue to exploit the opportunities that the AWS infrastructure provides by rolling out further mirrors over the course of 2011.
Increased functionality in the core Ensembl code base is at the heart of our efforts to support new features and continue to scale to the growing size of our data resources. For example, in order to accommodate the computational requirements of our data production process, the Ensembl API has been extended to support simultaneous access to databases on separate servers. We have also introduced a ‘production’ database to organize information collected throughout the release process. The production database consolidates and extends information that was previously held in a large number of disparate files.
Ensembl outreach
Ensembl’s commitment to user support, outreach and training helps us reach out to new communities and identify emerging trends in use of the project. For example, an analysis of more than 1500 queries received by the helpdesk from users in the past year reveals a growing trend towards data retrieval, either through programmatic access to our databases or through BioMart. Interest in SNPs and other genomic variation also ranks increasingly high with users. There has been a steady increase in training activities with 99 events held in 26 different countries. We have been reaching out for new communities showing an interest in genomics such as research clinicians, including four workshops for the UK National Health Service (NHS) in 2010.
We continue to improve Internet-based methods to communicate with users, such as: the introduction of dynamic ranking of Frequently Asked Questions (FAQs) based on user feedback; our YouTube channel featuring 12 training videos accessed over 30 000 times; and our blog and twitter feeds. These new methods expand and complement our efforts to introduce Ensembl through more traditionally published tutorials (15).
FUNDING
The Wellcome Trust; European Union (partial); the UK Biotechnology and Biological Sciences Research Council (partial); the National Human Genome Research Institute of the US National Institutes of Health (partial); the European Molecular Biology Laboratory (partial). Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all of our users and especially those who contacted us with feedback through our mailing list and during Ensembl courses. We acknowledge those researchers, organizations and large-scale projects that have provided data to Ensembl prior to publication under the understandings of the Fort Lauderdale meeting discussing Community Resource Projects and the Toronto meeting on prepublication data sharing.
REFERENCES
- 1.Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foelo ML, Sherry ST. NCBI dbSNP Database: content and searching. In: Weiner MP, Gabriel SB, Stephens JC, editors. Genetic Variation: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 41–61. [Google Scholar]
- 4.UniProt Consortium. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC genome browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Künstner A, Searle S, White S, Vilella AJ, Fairley S, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Bloomberg L, Bouffard P, Burt DW, Crasta O, Crooijmans RP, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 8(9):e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stabenau A, McVicker G, Melsopp C, Proctor G, Clamp M, Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker A, Bragin E, Brent S, Pritchard B, Smith JA, Trevanion S. Using caching and optimization techniques to improve performance of the Ensembl website. BMC Bioinformatics. 2010;11:239. doi: 10.1186/1471-2105-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rios D, McLaren WM, Chen Y, Birney E, Stabenau A, Flicek P, Cunningham F. A database and API for variation, dense genotyping and resequencing data. BMC Bioinformatics. 2010;11:238. doi: 10.1186/1471-2105-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severin J, Beal K, Vilella AJ, Fitzgerald S, Schuster M, Gordon L, Ureta-Vidal A, Flicek P, Herrero J. eHive: an artificial intelligence workflow system for genomic analysis. BMC Bioinformatics. 2010;11:240. doi: 10.1186/1471-2105-11-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spudich GM, Fernández-Suárez XM. Touring Ensembl: a practical guide to genome browsing. BMC Genomics. 2010;11:295. doi: 10.1186/1471-2164-11-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Cunningham F, Rios D, McLaren WM, Smith J, Pritchard B, Spudich GM, Brent S, Kulesha E, Marin-Garcia P, et al. Ensembl variation resources. BMC Genomics. 2010;11:293. doi: 10.1186/1471-2164-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballester B, Johnson N, Proctor G, Flicek P. Consistent annotation of gene expression arrays. BMC Genomics. 2010;11:294. doi: 10.1186/1471-2164-11-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper DN, Stenson PD, Chuzhanova NA. The Human Gene Mutation Database (HGMD) and its exploitation in the study of mutational mechanisms. Curr. Protoc. Bioinform. 2006;21:1.13.1–1.13.20. doi: 10.1002/0471250953.bi0113s12. [DOI] [PubMed] [Google Scholar]
- 20.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalgleish R, Flicek P, Cunningham F, Astashyn A, Tully RE, Proctor G, Chen Y, McLaren WM, Larsson P, Vaughan BW, et al. Locus reference genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, DiCuccio M, Yaschenko E, et al. Public data archives for genomic structural variation. Nat. Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–1828. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paten B, Herrero J, Fitzgerald S, Beal K, Flicek P, Holmes I, Birney E. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18:1829–1843. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paten B, Herrero J, Beal K, Birney E. Sequence progressive alignment, a framework for practical large-scale probabilistic consistency alignment. Bioinformatics. 2009;25:295–301. doi: 10.1093/bioinformatics/btn630. [DOI] [PubMed] [Google Scholar]
- 31.Cooper GM, Stone EA, Asimenos G, NISC Comparative Sequencing Program. Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]