Abstract
Protein–protein interactions are central to almost any cellular process. Although typically protein interfaces are large, it is well established that only a relatively small region, the so-called ‘hot spot’, contributes the most to the total binding energy. There is a clear interest in identifying hot spots because of its application in drug discovery and protein design. Presaging Critical Residues in Protein Interfaces Database (PCRPi-DB) is a public repository that archives computationally annotated hot spots in protein complexes for which the 3D structure is known. Hot spots have been annotated using a new and highly accurate computational method developed in the lab. PCRPi-DB is freely available to the scientific community at http://www.bioinsilico.org/PCRPIDB. Besides browsing and querying the contents of the database, extensive documentation and links to relevant on-line resources and contents are available to users. PCRPi-DB is updated on a weekly basis.
INTRODUCTION
Proteins are highly sociable molecules. The reason is that proteins catalyze complex biochemical reactions and are responsible of coordinating intricate cellular tasks; therefore they act as highly coordinated complexes rather than as isolated entities. Indeed, protein–protein interactions (PPIs) underlie most of the reactions that take place in cells and make life possible. Of particular interest in the study of PPIs is the description of the so-called ‘hot spot(s)’ of the interaction. The concept of hot spot originates from the seminal work by Clackson and Wells (1) (and subsequent research) which proved that most of the binding energy associated to a given PPi can be ascribed to a small set of complementary interface residues that contribute the most to the binding energy, i.e. the hot spot of the interaction.
The study and identification of hot spots in protein interfaces is an important and relevant question that has clear applications in drug discovery (2) and protein design. However, experimental techniques including Alanine scanning (3), Alanine shaving (4) or residue grafting (4), are lengthy, labor intensive and costly. Computational tools, such as our recently described Presaging Critical Residues in Protein interfaces (PCRPi) method (5) can be used to assist and complement experimental efforts. Under benchmark conditions, PCRPi delivered highly consistent and accurate predictions of hot spot residues in protein interfaces (5), thus justifying its use as predictive tool. Here, we present Presaging Critical Residues in Protein interfaces Database (PCRPi-DB), the result of the annotation and archiving of the entire Protein DataBank (6) (PDB) using PCRPi.
PCRPi-DB is a public repository of computationally annotated hot spots in protein complexes for which the 3D structure is known. The updating process is fully automated resulting on PCRPi-DB being updated once a week when new protein structures are released in the PDB. To date PCRPi-DB archives 68 589 protein structures (176 719 protein chains), of which 90 475 protein chains have been annotated, amounting to 4 844 157 interface residues. PCRPi-DB features a clear and intuitive web interface that allows users to search and retrieve data easily and conveniently. Furthermore, PCRPi-DB is cross-linked to several major databases thus increasing the range of information offered to users.
ANNOTATION, IMPLEMENTATION, CONTENTS AND USE
Prediction algorithm: PCRPi
Interface residues that are located in a hot spot present certain characteristics that are specific to them. Those have been exploited for predictive purposes including energy (i.e. in silico Alanine scanning), structure (e.g. solvent accessibility) and evolutionary-based (e.g. sequence conservation) features. Although these descriptors are useful, it was shown that individually they cannot unambiguously define hot spots (7). PCRPi (5) overcomes this limitation by combining a set of seven different measures that account for energetic, structural and evolutionary information into a common probabilistic framework by using Bayesian Networks (BNs) (8). PCRPi was benchmarked in two independent datasets and under both scenarios PCRPi delivered highly accurate and consistent predictions. Moreover, in a head-to-head comparison with other available computational tools using the same test set, PCRPi predictions were superior in terms of precision, recall and F1-scores (5).
PCRPi features two types of BNs: a naive and an expert, which can be trained in two different data sets: Ab+ and Ab−. Naive BNs assume that measures are independent whereas expert BNs allow conditional dependence between input measures. The difference between Ab+ and Ab− training sets is that Ab+ training set includes non-evolutionary related complexes such antigen–antibody complexes. The distinction was made due to the fact that antigen–antibody complexes do not have a common evolutionary history and therefore evolutionary-based measures are of no use. More information about the structure of the BNs and the composition of the training sets can be found in the help pages of the server or in the original publication describing the method (5). Therefore, each interface residue can be characterized by four different probabilities depending of the type of BN and the training set used during prediction (see ‘Annotated data’ section).
Database implementation
PCRPI-DB comprises two major components: a relational database management system for data storage and management and a web application to interface the database. Data are stored in a relational MySQL database whose design was optimized to provide a fast and optimal access to the information. It makes extensive use of master and internal keys and cross-references between tables. The MySQL server runs in a dedicated computer that also mirrors all external databases that are required during the updating and annotation process, e.g. PDB (6).
The web application runs on an Apache webserver hosted on a Red Hat® enterprise Linux operating system. CGI Perl, JavaScript and DBI-DBD modules are used to interface and access the database. Web pages resulting from queries and sequence searches are generated ‘on the fly’, i.e. are dynamic, thus ensuring up-to-date information is available to users. The website includes a Jmol applet to visualize protein structures and a BLAST (9) search engine to perform sequences searches (see ‘Retrieving information’ section). As a general rule, table headers, icons and other elements shown in web pages are active, i.e. hovering over them will reveal a short help and/or perform a task (e.g. reveal a table). Besides, there is an extensive documentation and explanations about contents, annotation process and the use of PCRPi-DB in the help pages.
Database contents
As explained, hot spots are annotated using PCRPi (5). PCRPi requires the atomic coordinates of the protein complex in standard PDB format, thus in principle any protein complex deposited in the PDB databank would be annotated in PCRPi-DB. However, annotations are restricted to protein complexes solved by X-ray crystallography with a crystal resolution better than 3.0 Ang. and therefore, NMR, structural models, protein–non-protein complexes (e.g. protein–DNA), single-chain protein structures or multi-chain protein structures that lack inter-chain atomic interactions and X-ray structures solved at a resolution worse than 3.0 Ang. are not included in PCRPi-DB. Also, proteins are filtered by size (i.e. number of residues) and protein chains shorter than 50 residues are not considered.
Prior to annotation, protein structures undergo a set of checks. The atomic coordinates of non-standard amino acids [with the exception of selenomethione (MSE) that is converted into methionine (MET)], and non-protein molecules (e.g. DNA) are discarded. If atoms present alternative locations, then only the first location or rotamer is kept. Also, residues having insertion codes are structurally superimposed and discarded if structurally equivalent. Missing main- and side-chain atoms are added using Maxsprout (10) and Scwrl 4.0 (11), respectively. All these steps ensure the quality of protein structures and minimize the errors associated to the computational estimation of changes in binding energy (i.e. in silico Alanine scanning) that are highly affected by the quality of the structure (e.g. missing atoms).
The second set of checks implies the comparison between asymmetric (ASU) and biological (BIOU) units. ASUs represent the smallest unit of the crystal whereas BIOUs are believed to represent the functional assembly of proteins in vivo. Usually ASU and BIOU are similar but they can differ. Differences include: (i) protein is known to act as a monomer but crystallize in mutimeric form; (ii) although protein acts as a multimer, the multimeric state reported by the ASU is not correct; crystallographic symmetry operations (i.e. rotations and translations) are required in order to generate the correct assembly or (iii) ASU only represents part of the BIOU, and thus requiring crystallographic symmetry operations of all or parts of the ASU. In all these situations, ASUs cannot be used because interfaces are either false (first two cases; and thus not included in PCRPi-DB) or missing (second and third case). Instead of using ASUs, interfaces are extracted from BIOUs that are generated using the crystallographic symmetry operations reported in the header (REMARK 350) of the PDB file. Interfaces extracted from BIOUs and are annotated as: ‘Interface(s) extracted from biounits’ in PCRPi-DB.
PCRPi-DB is updated on a weekly basis after new protein structures are released in the PDB databank (usually Friday night). The NCBI reference sequences (RefSeq) database (12), used during the annotation process to cull homologous sequences to derive sequence profiles, is also weekly updated prior to annotation. The entire update process is fully automated and predictions are submitted to a computer farm, therefore the entire annotation process and upload of new data to the MySQL server is done within few hours after the release of new protein structures. Up-to-date information about database contents and date of last update is presented in the home page.
Retrieving information
There are two basic approaches to query and retrieve annotated data from PCRPi-DB. The first approach is by simply providing the PDB identification code of the protein complex of interest in the text box embedded in the top menu (Figure 1A). The server will return a web page containing general information and annotated hot spots related to the given PDB identification code (see ‘Annotated data’ section). The second approach is by doing a sequence search using a BLAST (9) engine implemented in the web server (Figure 1B). In this case, users should enter or upload the protein sequence (raw or FASTA format only) and, if required, an E-value, cut-off value and substitution matrix can be selected in the advanced option menu (Figure 1B). The server will return a list of target proteins sorted by the E-value. Users can inspect the BLAST alignments by clicking on the relevant links where interface residues are highlighted in red (Figure 1C). The list also contains the links to each individual protein chain (see ‘Annotated data’ section).
Figure 1.
Querying PCRPi-DB. (A) Query using the PDB identification code. (B) Searching for homologous proteins using BLAST. (C) Detail of a BLAST alignment where interface residues are highlighted in red.
PCRPi-DB also features an advanced search engine that allows more complex and elaborated queries. Users can query the database by any of the following methods: (i) search for protein complexes that have an interface surface area smaller/equal/larger than a selected cut-off (Å2); (ii) search for protein complexes that have less/equal/more interface residues than a selected cut-off; (iii) search for protein complexes that have less/equal/more annotated hot spot residues at a given probability cut-off; (iv) searches using free text or keywords (i.e. reductases), Unitprot accession number (13) or PubMed identifier; and (v) any combination of aforementioned queries.
Annotated data
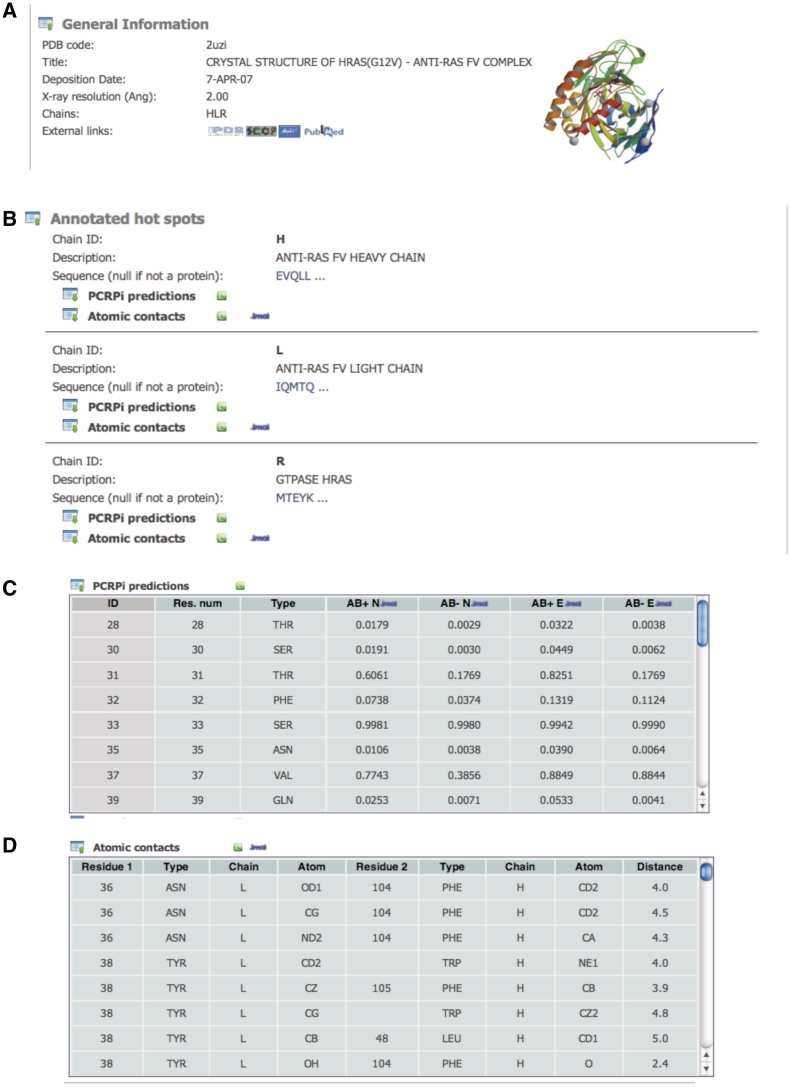
Each protein complex included in PCRPi-DB is presented in an individual web page that consists of two main expandable/collapsible sections: (i) a section that provides general information about the protein complex: ‘General information’; and (ii) the ‘Annotated hot spots’ section, which provides information about annotated hot spots and atomic interactions between protein chains.
The ‘General information’ section (Figure 2A) provides a quick overview and basic information of protein complexes including the PDB identification code, a brief description, the date when the coordinates were deposited in the PDB (6), X-ray crystal resolution, the number of chains and links to external resources, i.e. the PDB (6), SCOP (14) and Uniprot (13) databases, the digital object identifier (DOI) annotation system and the Pubmed database. Having easy and convenient access to relevant databases significantly expand the scope of the information that is available to the user. Under the ‘annotated hot spots’ section, protein chains are presented sequentially in an expandable/collapsible frame including the chain identification code, a brief description, protein sequence and two tables: PCRPi predictions and Atomic contacts (Figure 2B).
Figure 2.
Screenshots showing the information that is available for each of the protein complexes annotated in PCRPi-DB. (A) Detail of the ‘General information’ section. (B) ‘Annotated hot spot’ section. (C) Detail of the table showing the prediction of hot spot residues. (D) Detail of the table detailing atomic interactions between interface residues.
The PCRPi predictions table (Figure 2C) contains the prediction of hot spot residues and is composed of seven columns. The headers of the columns are active, thus hovering over them will reveal a short description about the contents. From left to right, first column is an internal residue identification number. The internal identification code is the residue number (different from the residue number shown in column 2 that corresponds to the residue number as in the coordinate file deposited in PDB databank), which is unique and is used to deal with cases when coordinates files contain insertion codes, rotamers, etc. In any case, hovering over the internal ID column will highlight the specific residue in the protein sequence. Column three shows the residue type in three-letters code and column four (AB+N), five (AB−N), six (AB+E) and seven (AB−E) refer to the prediction probabilities using a naive BN trained with the Ab+ dataset, a naive BN trained with the Ab– data set, an expert BN trained with the Ab+ data set and a naive BN trained with the Ab– data set, respectively (see ‘Prediction algorithm: PCRPi’ section). A link to download the data presented in the table in text tab-delimited plain format is provided along with a link to Jmol applets that allows the visualization of the prediction probabilities mapped onto the structure and some other manipulations (Supplementary Figure S1).
The atomic contacts table (Figure 2D) is composed of nine columns and provides information of non-bonded atomic interactions between interface residues as defined by the CSU program (15). Column headings are self-explanatory but hovering over them will show a short help description. The data contained in the table are downloadable in tab-delimited plain text format using the link provided, and atomic interactions can be visualized in the context of protein structure by using a Jmol applet (Supplementary Figure 2).
An example of an annotated protein complex: TEM-1 β-lactamase–β-lactamase inhibitor protein
β-Lactamases are enzymes that hydrolyze β-lactam bonds and thus confer resistance to β-lactam antibiotics like penicillins and cephalosporins to bacteria. The β-lactamase inhibitor protein (BLIP) is a natural inhibitor synthesized by species of the Streptomyces genus that binds to TEM-1 with subnanomolar affinity (16). The interface between TEM-1 and BLIP has been subjected to an extensive mutational analysis in order to discern the contribution of residues to the interaction (17). Four mutations in the BLIP interface: D49A, K74A, F142A and Y143A [residue numbering as in PDB code 1jtg (16)], resulted in a change in binding free energy of 7.5, 14.9, 8.8 and 1.6 kJ.mol−1, respectively. Therefore, three out of the four residues can be considered as critical or hot spot residues.
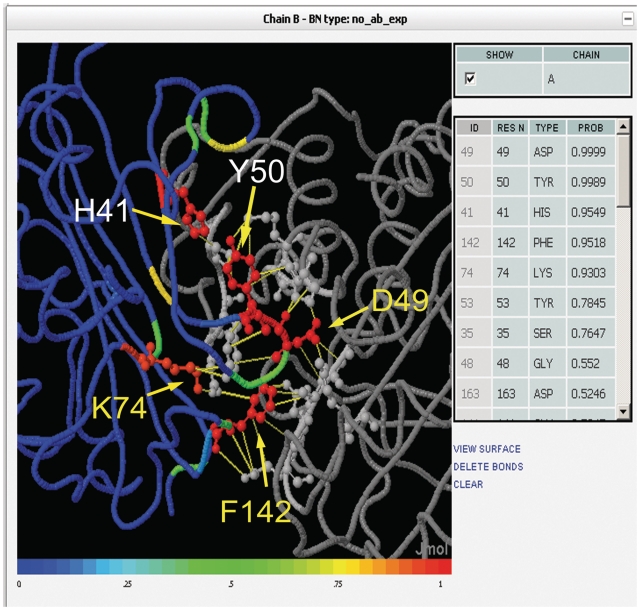
Comparing to the annotated data in PCRPi-DB for the given complex, PCRPi assigns probabilities higher than 0.9 to D49, K74 and F142, i.e. very likely to be critical to the interaction (Figure 3). Likewise, Y143 has a very low probability: 0.15; therefore, predictions fully agree with experimental observations. In addition, PCRPi assigns high probabilities to H41 and Y50 (Figure 3). Both residues are structurally close to K74, F142 and Y143 and may also be playing an important role in TEM-1/BLIP interaction. There are no experimental reports to confirm these predictions; however it illustrates one of the potential uses of the information contained in the database as guiding tool to pursue further experimental analysis, i.e. concentrate efforts in a subset of interface residues instead of a comprehensive exploration of the entire interface.
Figure 3.
Screenshot of a Jmol applet showing annotated hot spot residues on the TEM-1 β-lactamase–β-lactamase inhibitor protein (BLIP) complex [PDB code 1jtg (16)]. TEM-1 β-lactamase is depicted in gray and ribbon representation and BLIP is colored according to the predicted probability and shown in ribbon. Experimentally verified hot spot residues are labeled in yellow and presented in ball-and-stick representation.
CONCLUSION
A database of computationally annotated hot spots in protein interfaces, PCRPi-DB, is presented. The information available in PCRPi-DB has clear applications in drug discovery, structure-based protein design and can be also used in large-scale studies aimed at gaining further understanding on protein–protein interactions. PCRPi-DB has a clear and intuitive web interface and a number of functionalities that allow an easy and convenient access to the data. PCRPi-DB is weekly updated coinciding with the release of new protein structures in the PDB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Councils United Kingdom Academic Fellow scheme (to N.F.F.) and an internal scholarship awarded by the Leeds Institute of Molecular Medicine (to J.S.M.). Funding for open access: RUCK Academic Fellowship scheme.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
N.F.F. thanks Dr. Gendra for critical reading and insightful comments to the manuscript, and Ms Martina and Ms Daniela G. Fernandez for continuing inspiration and motivation. The authors acknowledge preliminary work done by Dr Assi in the implementation of the database. N.F.F. acknowledges constructive and insightful comments from anonymous referees.
REFERENCES
- 1.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 2.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 3.Wells JA. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 4.Jin L, Wells JA. Dissecting the energetics of an antibody-antigen interface by alanine shaving and molecular grafting. Protein Sci. 1994;3:2351–2357. doi: 10.1002/pro.5560031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assi SA, Tanaka T, Rabbitts TH, Fernandez-Fuentes N. PCRPi: Presaging Critical Residues in Protein interfaces, a new computational tool to chart hot spots in protein interfaces. Nucleic Acids Res. 2009;38:e86. doi: 10.1093/nar/gkp1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 7.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 8.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. San Francisco: Morgan Kaufmann Publisher Inc; 1988. [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Holm L, Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J. Mol. Biol. 1991;218:183. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 11.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard TJ, Murzin AG, Brenner SE, Chothia C. SCOP: a structural classification of proteins database. Nucleic Acids Res. 1997;25:236. doi: 10.1093/nar/25.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 16.Lim D, Park HU, De Castro L, Kang SG, Lee HS, Jensen S, Lee KJ, Strynadka NC. Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase. Nat. Struct. Biol. 2001;8:848–852. doi: 10.1038/nsb1001-848. [DOI] [PubMed] [Google Scholar]
- 17.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc. Natl Acad. Sci. USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]