Abstract
The RNA-Binding Protein DataBase (RBPDB) is a collection of experimental observations of RNA-binding sites, both in vitro and in vivo, manually curated from primary literature. To build RBPDB, we performed a literature search for experimental binding data for all RNA-binding proteins (RBPs) with known RNA-binding domains in four metazoan species (human, mouse, fly and worm). In total, RPBDB contains binding data on 272 RBPs, including 71 that have motifs in position weight matrix format, and 36 sets of sequences of in vivo-bound transcripts from immunoprecipitation experiments. The database is accessible by a web interface which allows browsing by domain or by organism, searching and export of records, and bulk data downloads. Users can also use RBPDB to scan sequences for RBP-binding sites. RBPDB is freely available, without registration at http://rbpdb.ccbr.utoronto.ca/.
INTRODUCTION
RNA-binding proteins (RBPs) have a fundamental role in a wide variety of cellular processes including transcription, RNA splicing and processing, localization, stability and translation (1–6). RBPs typically contain RNA-binding domains (RBDs) such as the RNA Recognition Motif (RRM) and the K homology (KH) domain, which are among the most numerous protein domains in metazoan genomes, including the human genome (7–9). Individual RBPs often have multiple RBDs that can independently bind RNA (10), and the approximately 400 annotated mammalian RBPs contain over 800 individual RBDs (11).
Knowledge of the RNA-binding activity of RBPs is critical for mapping and understanding transcriptional and post-transcriptional networks and regulatory mechanisms. Collections of DNA-binding specificities of transcription factors are available and widely used (12,13); however, to our knowledge, there is no central repository of information on the RNA-binding activities of RBPs. Here, we introduce RNA-Binding Protein DataBase (RBPDB), a database of RNA-binding experiments. A total of 1453 in vitro and in vivo experiments on 272 proteins are included, as well as 71 binding profiles in the form of position weight matrices (PWMs) and sequence logos, and 36 sets of sequences bound in vivo in immunoprecipitation experiments.
We anticipate that RBPDB will be of use to diverse researchers. In addition to searching for RNA-binding activities by protein, domain and experiment, RBPDB also allows users to scan RNA sequences for matches to RBP binding preferences stored in RBPDB. Additionally, the collected motifs should prove invaluable for genome-wide scans to identify cis-regulatory elements involved in post-transcriptional regulation via RBPs. Finally, the inclusion of in vivo bound transcripts provides a snapshot of enriched RBP-specific mRNA targets.
DATABASE DESIGN AND IMPLEMENTATION
Overview
RBPDB is a collection of RBPs linked to a curated database of published observations of RNA binding. The database consists of a table of proteins, linked to other proteins through orthology relationships and to one or more experiments, if experiments are found. Each protein and experiment is assigned a unique internal ID number, and proteins are linked to Ensembl, FlyBase and WormBase gene annotations and RNA-bound protein structures on PDB (14–17). Experiments are associated with a PubMed ID. Motifs, PWMs and large-scale data sets are retained as flat files that are linked to experiment and protein IDs.
Protein catalog
To populate the database, we first cataloged known and predicted RBPs in human, mouse, Drosophila and Caenorhabditis elegans (18–26). Most proteins were selected based on the presence of known sequence-specific RBDs (Table 1), which we compiled from review papers (3,4,7,8) and from searching and scanning Pfam domain annotations (27). We retrieved protein matches to InterPro domains from UniProt and Ensembl and used the union of these two sets. Additionally, we added proteins that bind RNA through a non-canonical RBD, such as a Sterile Alpha Motif (SAM) domain or C2H2 zinc finger, based on a Gene Ontology or keyword annotation as RNA-binding in Ensembl, UniProt or NCBI. However, we did not include domains that are largely specific to ribosomal proteins (e.g. S4 domain). Moreover, some non-sequence specific, poorly characterized and/or unconventional RBDs are currently not included (e.g. dsRBD, G-patch, zinc-knuckle and zinc-ribbon) (7). Inclusion of additional domains and species is a future objective for RBPDB, and users can suggest novel domains for inclusion (see Future Directions section). We note, however, that in eukaryotes, the repertory of known and predicted RBPs is dominated by RRM and KH domains, and as such, these constitute the majority of experimental data in RBPDB.
Table 1.
Current species and protein domain coverage in RBPDB
| Species | Number of proteins |
| Human | 422 |
| Mouse | 413 |
| Fly (Drosophila melanogaster) | 258 |
| Worm (Caenorhabditis elegans) | 244 |
| RNA-binding domain | Number of proteinsa |
| RNA Recognition Motif | 733 |
| CCCH zinc finger | 225 |
| K Homology | 138 |
| Like-Sm domain | 81 |
| C2H2 zinc finger | 30 |
| Ribosomal protein S1-like | 32 |
| Cold-shock domain | 29 |
| Lupus La RNA-binding domain | 26 |
| Pumilio-like repeat | 23 |
| Pseudouridine synthase and archaeosine transglycosylase (PUA domain) | 21 |
| Surp module/SWAP | 19 |
| Sterile Alpha Motif | 11 |
| YTH domain | 12 |
| PWI domain | 10 |
| THUMP domain | 9 |
| TROVE module | 6 |
aMany proteins have more than one RBD.
A short text description of the RBDs in the largest isoform of the protein (e.g. RRMx2 for a protein with two RRM domains) was assigned, and links to UniProt were added where available. In addition, in order to facilitate comparison between the RNA-binding specificities of similar proteins in different organisms, we imported orthology relationships from InParanoid (28).
During the course of curation, when we encountered RNA-binding experiments for proteins in other species (such as Xenopus, yeast or rat), we added them to the database on an ad hoc basis. However, coverage of the RNA-binding proteomes of species other than human, mouse, Drosophila and C. elegans is not intended to be comprehensive.
Types and representation of RNA protein interactions
We populated RBPDB with RNA-binding data by searching PubMed with the gene names and aliases of the aforementioned RBPs, and recording any RNA-binding data found in the retrieved papers. RBPDB currently catalogs 14 types of RNA-binding experiments. These include experiments that measure binding to a single sequence and those that measure binding to many sequences in parallel, in vivo or in vitro. A description of the categories of experiments and the number of experiments in each category is given in Table 2.
Table 2.
Types and numbers of experiments currently contained in RBPDB
| Experiment type | Description | Number of experiments in RBPDB |
|---|---|---|
| EMSA | Electromobility shift assays measure binding to a single RNA sequence in vitro by observing a change in RNA migration rate caused by binding to protein. | 522 |
| UV cross-linking | A single radiolabeled RNA sequence is cross-linked in cellular extract using UV radiation, and the bound proteins are separated by gel electrophoresis. Protein identity is determined using mass spectrometry or a protein-specific antibody. | 234 |
| Protein affinity purification | A synthetic RNA oligo or in vitro transcribed RNA is derivatized with a functional group, usually biotin, which allows it to be immobilized on streptavitin beads or affinity column. Cellular extract is applied, and the proteins that bind to the RNA are identified using antibodies. | 156 |
| SELEX | High-affinity binding sequences are selected from a randomized pool by several sequential rounds of binding to purified protein and PCR amplification. The resulting RNAs are cloned and sequenced, providing a set of short sequences preferred by the protein, which are analyzed for motifs, consensus sequences and structural preferences. | 117 |
| Genome-wide RNA immunoprecipitation | These methods assay for cellular RNAs bound to a protein in vivo, and include RIP-chip (or RIP-seq) where RNA is purified by immunoprecipitation with an antibody to the protein (41); HITS-CLIP (or CLIP-seq), where the immunoprecipitation is preceded by UV cross-linking (CLIP) (42); and PAR-CLIP where cross-linked sites are marked by an induced thymidine to cytidine transition (43). Affinity tags and RNA fragmentation are used in some cases. RNAs are detected by microarray or sequencing. A short motif can be detected in some cases, especially if the detected RNA fragments are short and numerous. | 91 |
| Filter binding assay | A single radiolabeled RNA is incubated with protein and filtered through a nitrocellulose filter. Protein-bound RNA is retained and detected. | 73 |
| Homopolymer-binding assay | The protein is typically incubated with agarose beads bound to a homoribopolymer sequence. The preference of the protein for poly(A), poly(C), poly(G) or poly(U) can be determined. | 69 |
| NMR | Nuclear magnetic resonance spectroscopy can be used to determine nucleotide-amino-acid level interactions for RBPs. | 64 |
| Fluorescence methods | This category includes several methods of measuring binding of a protein to a single fluor-tagged RNA sequence. | 47 |
| Yeast three-hybrid assay | In the yeast three-hybrid system, a modification of the yeast two-hybrid system for measuring protein–protein interactions, binding to the RNA of interest is measured by transcription of a reporter gene in yeast. | 30 |
| Yeast three-hybrid screen | The yeast-three hybrid system is applied to a library of RNA sequences in parallel. | 12 |
| Biosensor analysis | A method of detecting interactions between biomolecules using an RNA molecule coupled to a piezoelectric crystal. Binding to the protein of interest is detected by surface plasmon resonance. | 10 |
| RNAcompete | In the RNAcompete assay, a pool of RNA designed for specific sequence and structural features is incubated in excess to a GST-tagged protein. RNAs compete to bind to the protein, and the relative enrichment in the pulldown versus the pool is determined by microarray (44). | 9 |
| Other | This category includes rare methods such as isothermal titration calorimetry, single RNA immunoprecipitation or affinity purification and enzymatic RNA footprinting. | 13 |
Single-sequence experiments
Single-sequence experiments were included where the sequence of the bound RNA could be determined and is less than 200 nt in length. For these experiments, the full nucleotide sequence is included, unless a consensus motif rather than a unique sequence is reported. The consensus sequences use IUPAC (International Union of Pure and Applied Chemistry) nomenclature for representing degenerate nucleotides. Additionally, sequences with variable-length stretches or repetitive motifs are reported as (M)(X), where M is the repeated nucleotide or sequence, and X is a numerical value/range or a long undefined sequence (denoted as ‘n’). For example, the motif CUCUCU(A)(15–30)CUCUCU described for PTB contains two CUCUCU sequences separated by 15–30 adenosines (29), while (G)(n) denotes a poly(G) sequence.
SELEX experiments
For SELEX experiments, we extracted the selected sequences from the publication and aligned them as reported. We then created a position frequency matrix (PFM) from the alignment, and calculated a PWM using the Transcription Factor Binding Site (TFBS) package (30). Logos were created using the WebLogo standalone package (31). Reported motifs that contained internal gaps that would preclude representation in matrix format, or those for which >10% of the selected sequences do not match the reported motif, are reported as an IUPAC consensus motif only, as described above.
Large-scale in vivo binding experiments
When possible, we compiled all sequences identified in large-scale in vivo binding experiments. There is considerable diversity in how these data and sequences are reported and annotated. In some cases, we were unable to recover sequences; in these cases, RBPDB refers to the original publication but does not contain the sequences. When we were able to recover bound sequences, we included a short README file to describe how the sequences were extracted from supplementary data or GEO (Gene Expression Omnibus) (32). In general, when bound sequences were detected by tiling arrays, we extracted genomic sequence from the sense strand with respect to the annotated gene located ±200 bp of all reported peaks, since it is possible that pre-mRNA is bound, along with any numerical value associated with the peak (e.g. log ratio intensity). When only the identity of bound genes or transcripts is reported, we compiled the transcript or gene sequence retrieved from GenBank using BioPerl (33), or from batch download files from FlyBase, and reported this sequence along with its associated numerical value. There were a variety of different normalization and reporting strategies reported in these studies, and wherever possible, we report only normalized data rather than raw data, but we capture any associated GEO or ArrayExpress (34) identifiers to allow users to access the data directly. When there are multiple samples or controls, we report each separately. In some cases, matrices or sequence logos were reported for genome wide in vivo immunoprecipitation experiments, and are included in the database.
Representation of RNA structural requirements
RBDs recognize specific RNA sequences, structures or both. RNA binding in vivo is presumably dependent on a combination of factors, including accessibility of the binding site (35) and interactions with cofactors (including other RBPs). A goal of RBPDB is to describe bound sequences with minimal interpretation, which conflicts with complications surrounding the representation and storage of RNA structure in a compact, unambiguous, computer- and human-readable format. For example, minimum free energy structures require a windowing function to select the region of RNA to fold and are too simple to represent suboptimal structures, which can be biologically functional. Therefore, in RBPDB we include only a yes/no indication of whether the original manuscripts discussed the secondary structure of the RNA. Users interested in predicting structure should consider the RNAfold webserver (among others) (36).
USING RBPDB
There are three main modes of interaction with RBPDB. The first is to search for RNA-binding experiments by RBP, by RBD, by species, by experiment type or by any combination of the above. The second is to perform bulk downloads of all RBPDB data or subsets of the data filtered in various ways. The third is to scan an input RNA sequence for potential binding sites for RBPs stored in RBPDB.
Searching for RNA-binding experiments
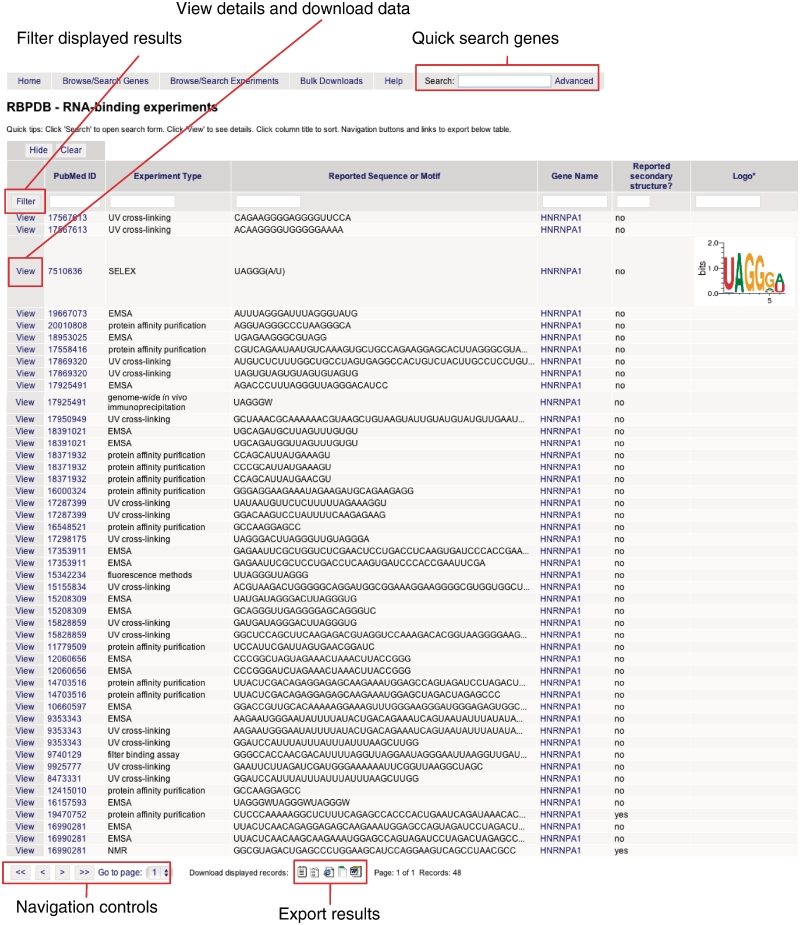
RBPDB can be searched quickly by gene name, alias or description, by entering a search term in the search box on the home page or at the top of every page. More complex queries can be executed using the advanced search form, reached by clicking the ‘advanced’ link. From here, the proteins database can be searched by gene name or symbol, organism, or RBDs by making the appropriate selections on the form. To retrieve experiment records directly, the experiments form should be used; it takes the same input, with the addition of options to search by experiment type. Figure 1 shows the results from one such search. From the results page, experimental data can be viewed and exported. Any results table can also be further filtered by partial text matches in any of the columns by clicking ‘Filter’. Columns can be sorted in decreasing or increasing order by clicking the column label.
Figure 1.
Example of searching RBPDB by gene name. Shown are results generated by using the advanced search form to search experiments. The query ‘HNRNPA1’ was entered in the gene name field and ‘human’ selected for species. Navigation links and links to view detailed information are indicated, as are the icons to export data in text, CSV, Excel, HTML and Word formats.
Bulk download of annotation, transcript and matrix data
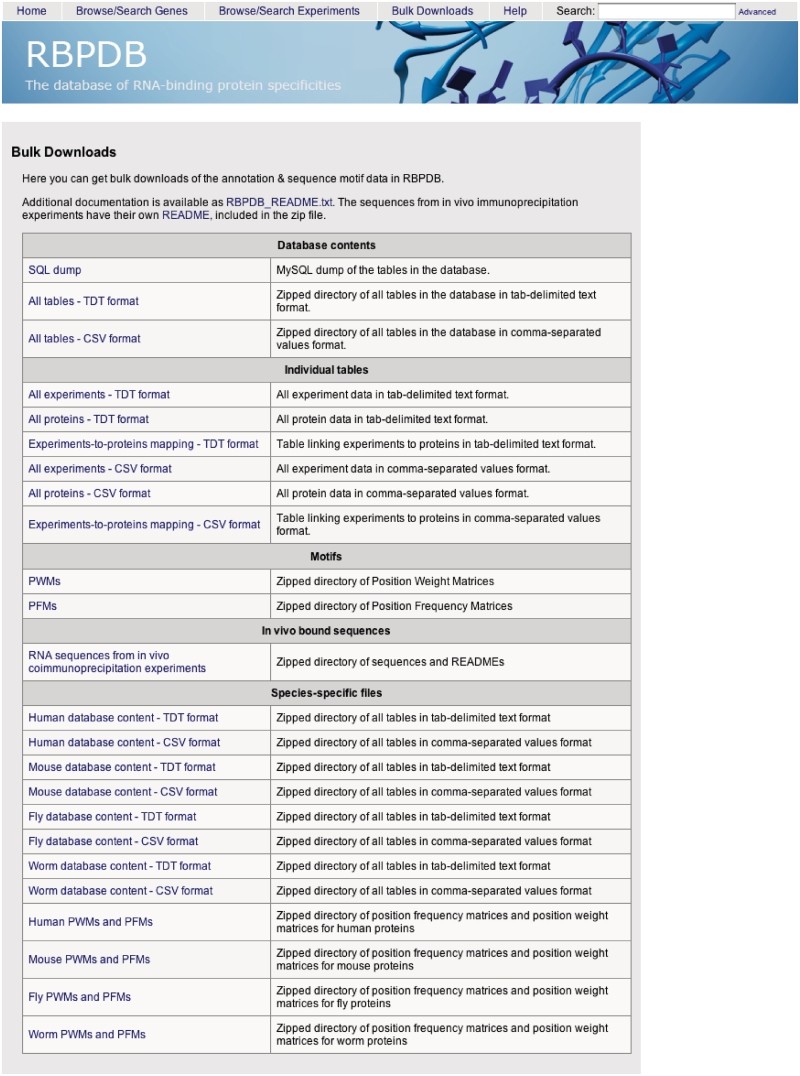
There are two ways to download data from RBPDB. First, the annotation data corresponding to a subset of proteins or experiments resulting from a search query can be exported in plain text, comma separated values (CSV), Excel or Word formats directly from a search result table, as shown in Figure 1. The second way to download data is via the Downloads page, linked from the menu at the top of the site (Figure 2). This page has links to files that include the full annotation database in SQL, tab-delimited and CSV formats, as well as sets of transcripts bound in genome wide in vivo experiments, and binding specificity PFM and PWM matrices in a flat text file format (30). The individual protein and experiment tables are also available, as well as the linker table needed to map experiments to proteins. These files are also available for each species separately.
Figure 2.
Download page of RBPDB. This screenshot shows the bulk data set downloads available.
Scanning input sequences for RBP-binding sites
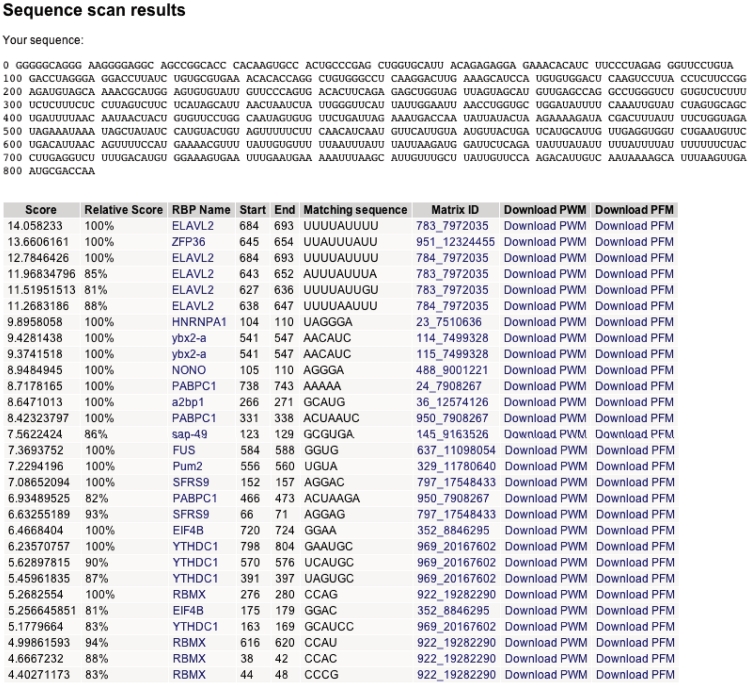
From the main page, users can submit nucleotide sequences to scan for matches with RBP-binding sites. This sequence can be in DNA or RNA format. Additionally, a threshold for reporting matches to the sequence can be set. At present, the sequence can only be scanned with motifs associated with full PWMs. Potential binding sites in the sequence are identified by scoring potential binding sites within the sequence using PWMs, using BioPerl (33). The PWM score for a potential binding site is the sum of the scores of each nucleotide at each position in the PWM, and the relative score is the percent of the score relative to the maximum possible score of the PWM calculated. Sites with relative scores greater than the threshold, which defaults to 80%, are reported. Figure 3 shows the results obtained for the 3′-UTR of the human c-fos gene. The RBPs TTP and members of the ELAV family have been implicated in the ARE-regulated degradation of c-fos RNA (37). The top hits are to known AU-rich element (ARE)-binding proteins ELAVL2 (HuB) and ZFP36 (TTP).
Figure 3.
Example of scanning input sequence for potential RBP-binding sites. The 3′-UTR of human c-fos was downloaded from GENBANK (Accession no. NM_005252, nucleotides 1349–2158) and submitted to the sequence scan form on the RBPDB home page.
It is also possible to search all individual RNA sequences from the single-sequence experiments by entering a sequence or IUPAC consensus of interest in the search window. The search will return exact matches to the text entered.
FUTURE DIRECTIONS
We will periodically update RBPDB to keep it current. Each protein entry in our database will be reassessed at least once a year. RBPDB also has a user submission form that allows users to notify our curators of recent publications of RNA-binding specificities or proteins newly discovered; we will prioritize these submissions for updates. Newly-described RBDs [e.g. the nudix domain (38)] and newly described RBPs without conserved domains will be included using the search strategy used for the initial construction of the database. A related future direction for RBPDB will be the systematic incorporation of data from other species. RBPDB is currently populated only with data from metazoans, which are of special interest for biomedical research, but represent only a small minority of the eukaryotic kingdom. There is RNA-binding information for proteins in other species, particularly traditional non-metazoan model systems such as yeast (39) and Arabidopsis [e.g. (40)], and also bacteria.
It may also be possible to further populate the database by inferring RNA-binding activities. While the existence of a universal molecular ‘code’ that predicts RNA sequence specificity directly from protein sequence has proven difficult to derive (25), there is little question that proteins with very similar amino-acid sequences tend to have very similar RNA-binding activities. As such, we anticipate that one application of RBPDB will be further analysis of the relationships between protein sequences and RNA-binding activities. For these analyses, it would be invaluable for the RNA-binding activities of individual RBDs to be documented, rather than individual proteins and the bound sequences to be aligned, if possible. Indeed, the way the RNA-binding activity is represented is critical for many uses of RBPDB, including genome scanning, identification of proteins that would bind sequences of interest, and comparisons among RBPs. Therefore, an area of ongoing exploration will be the representation of RNA-binding activities, including the inclusion of domain-specific information and incorporation of RNA structure.
FUNDING
Canadian Institutes of Health Research (MOP-93671 to T.R.H. and Q.M.; MOP-49451 to T.R.H.); National Institutes of Health (1R01HG00570 to T.R.H.); Natural Sciences and Engineering Research Council of Canada CGS-M (to K.C.). Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Harm van Bakel, Debashish Ray and Carl de Boer for computational support and helpful conversations.
REFERENCES
- 1.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKee AE, Silver PA. Systems perspectives on mRNA processing. Cell Res. 2007;17:581–590. doi: 10.1038/cr.2007.54. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Diaz P, Penalva LO. Post-transcription meets post-genomic: the saga of RNA binding proteins in a new era. RNA Biol. 2006;3:101–109. doi: 10.4161/rna.3.3.3373. [DOI] [PubMed] [Google Scholar]
- 4.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr. Opin. Cell Biol. 2008;20:144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 11.Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT, Mouse Genome Database G. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bult CJ, Blake JA, Richardson JE, Kadin JA, Eppig JT, Baldarelli RM, Barsanti K, Baya M, Beal JS, Boddy WJ, et al. The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res. 2004;32:D476–D481. doi: 10.1093/nar/gkh125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achsel T, Stark H, Luhrmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl Acad. Sci. USA. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- 21.Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Denhez F, Lafyatis R. Conservation of regulated alternative splicing and identification of functional domains in vertebrate homologs to the Drosophila splicing regulator, suppressor-of-white-apricot. J. Biol. Chem. 1994;269:16170–16179. [PubMed] [Google Scholar]
- 23.Aravind L, Koonin EV. THUMP–a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001;26:215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- 24.Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 2003;10:614–621. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- 25.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szymczyna BR, Bowman J, McCracken S, Pineda-Lucena A, Lu Y, Cox B, Lambermon M, Graveley BR, Arrowsmith CH, Blencowe BJ. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 2003;17:461–475. doi: 10.1101/gad.1060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berglund AC, Sjolund E, Ostlund G, Sonnhammer EL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamichhane R, Daubner GM, Thomas-Crusells J, Auweter SD, Manatschal C, Austin KS, Valniuk O, Allain FH, Rueda D. RNA looping by PTB: evidence using FRET and NMR spectroscopy for a role in splicing repression. Proc. Natl Acad. Sci. USA. 2010;107:4105–4110. doi: 10.1073/pnas.0907072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenhard B, Wasserman WW. TFBS: computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 31.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, Rustici G, Williams E, Parkinson H, Brazma A. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 2010;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 38.Yang Q, Gilmartin GM, Doublie S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc. Natl Acad. Sci. USA. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tam PP, Barrette-Ng IH, Simon DM, Tam MW, Ang AL, Muench DG. The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010;10:44. doi: 10.1186/1471-2229-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray D, Kazan H, Chan ET, Castillo LP, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]