Abstract
The ThYme (Thioester-active enzYme; http://www.enzyme.cbirc.iastate.edu) database has been constructed to bring together amino acid sequences and 3D (tertiary) structures of all the enzymes constituting the fatty acid synthesis and polyketide synthesis cycles. These enzymes are active on thioester-containing substrates, specifically those that are parts of the acyl-CoA synthase, acyl-CoA carboxylase, acyl transferase, ketoacyl synthase, ketoacyl reductase, hydroxyacyl dehydratase, enoyl reductase and thioesterase enzyme groups. These groups have been classified into families, members of which are similar in sequences, tertiary structures and catalytic mechanisms, implying common protein ancestry. ThYme is continually updated as sequences and tertiary structures become available.
INTRODUCTION
The ThYme (Thioester-active enzYme, http://www.enzyme.cbirc.iastate.edu) database presents enzymes acting on thioester-containing substrates, especially those involved in fatty acid and polyketide synthesis.
There are different ways to classify enzymes and proteins. The Enzyme Commission (EC) scheme classifies enzymes by the reactants or substrates that they primarily attack and by the reactions that they catalyze (1). Another way is by three-dimensional (tertiary) structure, as found in the SCOP database (2). A third method is to classify enzymes by primary (amino acid sequence) structure similarity. We have done so for thioesterases (TEs) (3) and now for the other enzyme groups in the fatty acid synthesis cycle. Previously, this has been done with glycoside hydrolases and other carbohydrate enzymes (4) and with peptidases (5). Also, Pfam (6) has done the same in a more universal way.
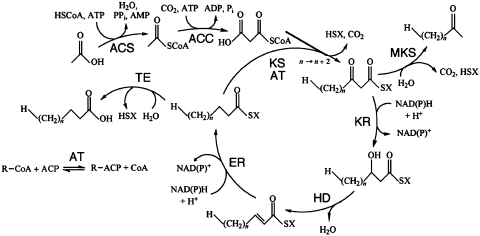
The fatty acid synthesis cycle (Figure 1) is the main pathway used by organisms to form lipids. The constituent members of this cycle are activated by the presence of thioester groups binding either coenzyme A (CoA) or acyl carrier protein (ACP). First, catalyzed by acyl-CoA synthases (ACSs), an acyl group is joined with CoA to make acyl-CoA, also called the priming substrate. Second, the priming substrate is carboxylated by acyl-CoA carboxylases (ACCs) to make the elongating substrate. The elongating substrate’s carrier molecule may be changed from CoA to ACP by acyl transferases (ATs). Then ketoacyl synthases (KSs) join the priming and elongating substrates, releasing a carbon dioxide and making ketoacyl-ACPs. The ketoacyl-ACP molecule then passes through a series of reduction, dehydration, and reduction steps catalyzed by ketoacyl reductases (KRs), hydroxyacyl dehydratases (HDs) and enoyl reductases (ERs), respectively, to create an acyl-ACP molecule two carbon atoms longer than the priming substrate. This new longer acyl-ACP molecule is then joined by a KS to another elongating substrate. This cycle elongates the acyl chain by two carbon atoms each turn until TEs hydrolyzes the CoA or ACP from the acyl group, effectively terminating fatty acid biosynthesis. Also, methylketone synthases (MKSs) can release molecules from the cycle before the reduction-dehydration-reduction steps. These enzymes first hydrolyze the thioester bond and then decarboxylate the carboxyl group of a 3-oxoacyl-ACP molecule, leaving a terminal methyloxo group (7). They have a TE domain, which appears in ThYme with other TEs; they do not form a large enzyme group.
Figure 1.
The fatty acid synthesis cycle and the enzyme groups that are part of it. ACC: acetyl-CoA carboxylase; ACS: acyl-CoA synthase; AT: acyl transferase; ER: enoyl reductase; HD: hydroxyacyl dehydratase; KR: ketoacyl reductase; KS: ketoacyl synthase; MKS: methylketone synthase; TE: thioesterase. SX: Coenzyme A or acyl carrier protein.
More specifically, the enzyme groups involved in the fatty acid synthesis cycle and that appear in ThYme are the following.
ACSs (part of EC 6.2.1, acid-thiol ligases). These enzymes add CoA to acetate or longer acceptors, powered by ATP or occasionally by GTP. This yields the activated compound and usually AMP, but in some cases ADP or GDP. ACSs are described by EC 6.2.1.1–EC 6.2.1.36, with two entries having been deleted.
ACCs (part of EC 6.4.1, ligases that form carbon-carbon bonds). In this step, the activated acceptor is elongated by the addition of a keto group derived from CO2, yielding malonyl-CoA or a longer activated molecule. Four multidomain ACCs with EC designations from 6.4.1.2 to 6.4.1.5 are listed.
ATs (part of EC 2.3.1, acyl transferases transferring groups other than amino-acyl groups). These enzymes catalyze the transfer of an acyl chain from a CoA to an ACP or vice versa.
KSs (part of EC 2.3.1, acyl transferases transferring groups other than amino-acyl groups). Here the activated malonyl or longer moiety is joined to an activated cycle constituent, releasing CO2 and HSX, where SX is CoA or ACP. The growing chain is elongated by generally two, but occasionally more, carbon atoms. This EC category contains 190 entries, of which three has been deleted. Twenty EC entries out of 187 are KSs.
KRs (part of EC 1.1.1, oxidoreductases acting on the CH–OH group of donors with NAD+ or NADP+ as acceptor, describing the reverse reaction). In those fatty acid synthesis cycle reactions, 3-oxo groups are reduced to 3-hydroxy groups by NADH or NADPH. EC 1.1.1. contains at present 300 entries, 15 having been deleted.
HDs (part of EC 4.2.1, carbon–oxygen hydro-lyases). Here the 3-hydroxy group is removed as water, yielding a double bond linking the 2- and 3-carbon atoms. There are 120 listings in this EC group, 16 having been deleted.
ERs (part of EC 1.3.1, oxidoreductases acting on the CH–CH group of donors with NAD+ or NADP+ as acceptor). The 2,3-ene bond is reduced to a single bond. This EC group has 84 listings, of which four have been deleted.
TEs (part of EC 3.1.2, thioester hydrolases). The thioester group is cleaved with water, leaving a fatty acid and HSX. The 27 EC entries have lost three members by deletion.
Polyketide biosynthesis is similar to fatty acid biosynthesis, yet it is more flexible and complex. Here the condensation-reduction-dehydration-reduction cycle is not completed at every turn; the KS-catalyzed reaction can occur between an intermediate in the cycle and an elongating substrate. This allows carbonyl, hydroxyl and/or ethylene groups into the acyl chain. The TE will either hydrolyze acyl-CoA or acyl-ACP with a water molecule, or cyclize the chain using an alcohol on the chain itself for hydrolysis. Also, different compounds can be used for priming and elongating substrates.
These processes can be carried out by individual independent enzymes, or by large multimodular fatty acid synthases (FASs) or polyketide synthases (PKSs) that contain the number of domains necessary, and in a specific order, to produce the desired molecule.
Among other uses, fatty acids have been recently proposed as biofuel feedstocks (8), while short-chain fatty acids could become feedstocks for biorenewable platform chemicals (9). Polyketides are a diverse family of chemicals, with some having medicinal applications such as erythromycin and tetracycline as antibiotics and doxorubicin and mithramycin in chemotherapy. Tailoring these molecules is of great interest; for that effort ThYme can be a useful tool in finding naturally occurring enzymes and in facilitating enzyme design.
IDENTIFYING AND POPULATING FAMILIES
Family members must have strong sequence similarity and near-identical tertiary structures, and they must share general mechanisms as well as catalytic residues located in the same position. Methods for identifying and populating families were developed with TEs and later applied to other sequence groups. They were detailed in our previous work and its Supporting Information section (3).
Experimentally confirmed enzyme sequences were used as queries. They were gathered from UniProt (10), using only reviewed entries noted as having ‘Evidence at protein level’.
A series of successive Basic Local Alignment Search Tool (BLAST) (11) searches and comparison among results reduced query sequences to a few representative ones.
The catalytic domains of representative query sequences were subjected to BLAST to populate the families. These domains were selected by referring to Pfam-A (6), or by constructing a hidden Markov model profile (12) from a multiple sequence alignment (MSA) based on the initial BLAST result.
Experimentally confirmed enzymes were surveyed to search for missing potential enzyme families.
The uniqueness of the families was confirmed by MSAs, by tertiary structure superposition and comparison, and by catalytic residue positions.
PRESENT CONTENT
At present, ACSs are divided into five families, ATs into one, KSs into five, KRs into four, HDs into six, ERs into six and TEs into 23. ACCs are multidomain proteins first shown as organized into domains followed by each domain divided into families: one family of the biotin carboxylase (BC) domain, one family of the biotin carboxyl carrier protein (BCCP), and two families of the carboxyl transferase (CT) domain appear. These enzyme groups’ annotation and sequences in each family appear in ThYme organized in the way mentioned below.
DATABASE ORGANIZATION AND FEATURES
The home page gives links to every enzyme group, as well as general information for viewers and citing and contact information. In each enzyme group’s main page, all families are listed in a table with ‘Names of enzymes and genes present’, which presents a non-exhaustive overview of the sequences found. This is meant to guide new users to the family that contains their enzymes of interest.
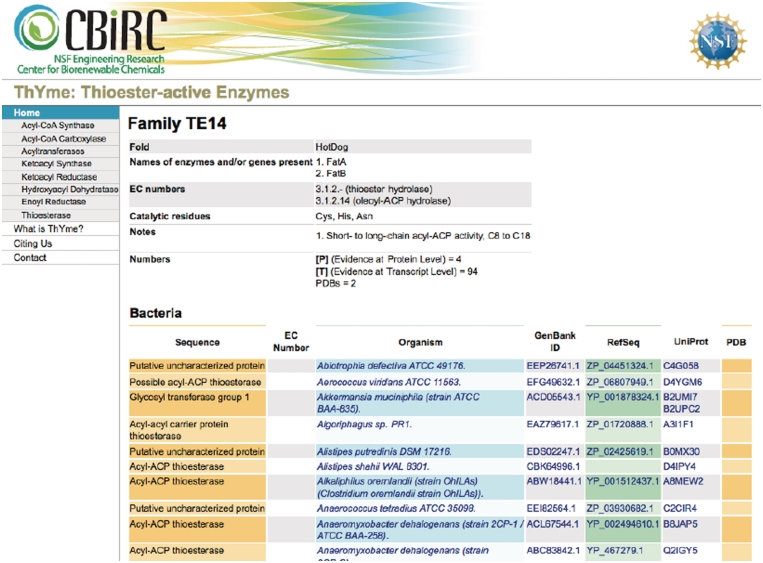
At the top of each enzyme family’s page (Figure 2), a table gives general information about the family, describing protein folds (if known from crystal structures), the names of enzymes and genes present (the list is not exhaustive), EC numbers (the most common ones), the catalytic residues (if they are known from the literature), and other notes. Also shown is the total number of Protein Data Bank (PDB) (13) structures, and enzymes with ‘Evidence at protein level’ and ‘Evidence at transcript level’ (see Experimentally Characterized sequences section below). This annotation might not be complete for all families.
Figure 2.
A typical family homepage in ThYme.
Within an enzyme family’s page, all sequences appear by rows ordered into archaea, bacteria and eukaryota, and alphabetically by producing species. All sequences in a row are identical and come from only one species. Identical sequences from different species are separated into different rows; however, identical sequences from different strains of the same species are not separated. If >500 rows exist, they are shown in multiple pages for a single family. The information is organized into the following columns: (i) names or designations given to the proteins; (ii) EC numbers assigned to them, with a link to the ExPASy proteomics server (14); (iii) genus and species names along with strain designations of the organisms that produced them, with a link to the National Center for Biotechnology Information (NCBI) taxonomy browser (15); (iv) their GenBank identification, with a link to the NCBI’s protein database (16); their RefSeq identification, with a link also to the NCBI’s protein database (16); their UniProt identification, with a link to the UniProt database (10); and their PDB identification, with a link to the PDB, if their known tertiary structure is available (13). All sequence names and EC numbers are taken from either UniProt or NCBI’s protein database; we do not assign sequence names or EC numbers.
Three features make navigating and retrieving information in ThYme easier. A search tool allows keywords, EC numbers and GenBank, RefSeq, UniProt or PDB accession codes to be searched. Furthermore, each family can be downloaded into a comma-separated value (csv) file, which can be viewed in a spreadsheet. Also, on each family’s page, only rows that include a PDB link or a UniProt link marked with ‘Evidence at transcript level’ or ‘Evidence at protein level’ can be viewed.
UPDATES
The content of existing families is updated continuously as NCBI’s protein database, UniProt and PDB databases are updated; if a new sequence belongs in an existing family, it will appear there. To delete or merge existing families, as well as to define new families, the authors’ inspection and judgment is necessary; this cannot be automated.
EXPERIMENTALLY CHARACTERIZED SEQUENCES
Most sequences have no underlying specific experimental work, as they come from large genomic sequencing projects. The UniProt database, under the field ‘Protein existence’ marks their entries with either ‘Evidence at protein level’ or ‘Evidence at transcript level’ if some experimental work has been done on the sequence. In ThYme, we mark UniProt accessions with ‘Evidence at Protein Level’ with a [P], and those with ‘Evidence at Transcript Level’ with a [T]. The UniProt link or its equivalent in GenBank shows the experimental work’s literature. This should help users identify previous work on enzymes of interest.
SEQUENCES WITH MULTIPLE DOMAINS
Some enzymes that appear in ThYme are multidomain FASs, PKSs or non-ribosomal peptide synthases. Each domain in these enzymes has its specific function, but all appear in a single sequence under the same GenBank, RefSeq, UniProt or PDB accession. When the accession code of a multidomain enzyme appears in a family, only the domain of the enzyme group in which the family appears belongs in the family. (Example: UniProt P12785 is a rat fatty acid synthase. Its AT domain appears in AT2, its KS domain appears in KS3, its HD domain appears in HD4 and its TE domain appears in TE16.) A single multidomain sequence can have different PDB structures for each domain. Only the structure related to each family’s domain is shown. (Example: UniProt P49327 has several PDB structures. Among them, TE domain 1XKT appears in a TE family, AT domain 2JFD appears in an AT family and so forth.)
SIMILARITY TO OTHER ENZYME DATABASES
ThYme is most similar to CAZy (17) in appearance and structure, in that both are interactive lists of enzyme primary and tertiary structures. However, they are different in content, as ThYme shows enzymes active on substrates with thioester groups and CAZy shows enzymes active on carbohydrates. ThYme encompasses eight enzyme groups; CAZy on the other hand brings together four enzyme groups as well as different families of carbohydrate-binding modules.
ThYme is somewhat similar to MEROPS (18), which classifies peptidases and therefore has many more different enzyme groups and total number of listings. MEROPS and ThYme are also different in appearance and in the method by which listings are accessed.
The ESTHER database (19) and the Lipase Engineering Database (20) report sequences of the α/β hydrolase superfamily and lipases, respectively. In both databases, some of their families correspond with some TE families in ThYme, although the exact content and format differ.
Finally, Pfam (6) has identified many protein families. Most ThYme families have an equivalent in Pfam. Our differences in methodology lead to different family content: Pfam families are more inclusive, covering a wide range of sequences, while ThYme families are smaller, with all sequences within a family having strong sequence similarity. Also, the purpose and format of the two databases are different; we focus on thioester-active enzymes and provide sequences and structures in families, while Pfam covers all proteins and, given a query, it identifies the family or domain.
CONCLUSION
The ThYme database should provide a useful source of information on these enzymes that can help predict active sites, catalytic residues and mechanisms of individual sequences, as well as providing a standardized nomenclature.
FUNDING
US National Science Foundation [through its Engineering Research Center Program, Award No. EEC-0813570, leading to the Center for Biorenewable Chemicals (CBiRC)], headquartered at Iowa State University and including Rice University, the University of California, Irvine, the University of New Mexico, the University of Virginia, and the University of Wisconsin–Madison. The authors are grateful for this support. Funding for open access charge: US National Science Foundation (through its Engineering Research Center Program, Award No. EEC-0813570).
Conflict of interest statement. None declared.
REFERENCES
- 1.Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) Enzyme Nomenclature. Recommendations 1992. Academic Press, San Diego. http://www.chem.qmul.ac.uk/iubmb/ [Google Scholar]
- 2.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 3.Cantu DC, Chen Y, Reilly PJ. Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrissat B. A classification of glycosyl hydrolases based in amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem. J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Israel I, Yu G, Austin MB, Bhuiyan N, Auldridge M, Nguyen T, Schauvinhold I, Noel JP, Pichersky E, Fridman E. Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 2009;151:1952–1964. doi: 10.1104/pp.109.146415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durrett TP, Benning C, Ohlrogge J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008;54:593–607. doi: 10.1111/j.1365-313X.2008.03442.x. [DOI] [PubMed] [Google Scholar]
- 9.Nikolau BJ, Perera MADN, Brachova L, Shanks B. Platform chemicals for a biorenewable chemical industry. Plant J. 2008;54:536–545. doi: 10.1111/j.1365-313X.2008.03484.x. [DOI] [PubMed] [Google Scholar]
- 10.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–W9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eddy SR. Profile hidden Markov models. Bioinformatics Rev. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 13.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hotelier T, Renault L, Cousin X, Negre V, Marchot P, Chatonnet A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Nucleic Acids Res. 2004;32:D145–D147. doi: 10.1093/nar/gkh141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer M, Pleiss J. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acids Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. [DOI] [PMC free article] [PubMed] [Google Scholar]