Abstract
Alternative splicing is emerging as a major mechanism for the expansion of the transcriptome and proteome diversity, particularly in human and other vertebrates. However, the proportion of alternative transcripts and proteins actually endowed with functional activity is currently highly debated. We present here a new release of ASPicDB which now provides a unique annotation resource of human protein variants generated by alternative splicing. A total of 256 939 protein variants from 17 191 multi-exon genes have been extensively annotated through state of the art machine learning tools providing information of the protein type (globular and transmembrane), localization, presence of PFAM domains, signal peptides, GPI-anchor propeptides, transmembrane and coiled-coil segments. Furthermore, full-length variants can be now specifically selected based on the annotation of CAGE-tags and polyA signal and/or polyA sites, marking transcription initiation and termination sites, respectively. The retrieval can be carried out at gene, transcript, exon, protein or splice site level allowing the selection of data sets fulfilling one or more features settled by the user. The retrieval interface also enables the selection of protein variants showing specific differences in the annotated features. ASPicDB is available at http://www.caspur.it/ASPicDB/.
INTRODUCTION
Alternative splicing is a well characterized mechanism which, coupled with alternative initiation and termination of transcription (1), may expand the transcriptome and proteome complexity in human and other organisms by over one order of magnitude with respect to the number of annotated genes (2,3). In particular, it is now widely demonstrated that virtually all multi-exon genes may generate multiple transcripts and protein variants (3,4) and that the splicing process is tightly regulated in different physiological conditions, tissues or developmental stages (5). Furthermore, alterations of the splicing process can be observed in several genetic diseases and in cancer (6–10).
The huge amount of EST sequences (11) together with the relevant reference genome sequence has been used to carry out an extensive analysis of alternative splicing in human through the ASPIC algorithm (12–14). The alternative splicing pattern of human multi-exon genes, determined by ASPIC, has been collected in ASPicDB, a database resource which presents some unique features with respect to other similar databases (15). The ASPIC algorithm implements an optimization strategy that, performing a multiple alignment of all available transcript data (including full-length cDNA and EST sequences) to the relevant genome sequence, detects the set of introns that minimizes the number of splicing sites. It also generates through a directed-acyclic graph combinatorial procedure the minimal set of non-mergeable transcript isoforms compatible with the detected splicing events (14). The reliability of splicing isoforms detected by ASPIC has been recently established through a comparative assessment (16).
The advent of massive transcriptome sequence data generated by RNA-Seq (17) is steadily increasing the number of validated splicing sites and isoforms in human and other organisms thus suggesting that a fraction of alternative splicing events are the result of background noise in the splicing process (18) which generates non-functional isoforms expressed at low level. Therefore, extensive research efforts are required to distinguish functional species-specific variants from non-functional ones originated from neutral drift in the splicing process, as well as to asses the biological role of functional isoforms.
The annotation of the protein variants predicted with ASPIC is an essential step for exploring the functional and structural diversity of the proteins originating from the same gene by means of alternative splicing and therefore for unraveling the complex physiological effects of alternative splicing events (19). Indeed, currently available databases, such as ASD (20), ASAP II (21), ASTALAVISTA (22) and H-DBAS (23), mostly collect information on alternative transcripts at the mRNA level, without considering the effect of alternative splicing on the protein structure and function. The ProSAS (24) database contains structural information as derived from comparative modeling procedure, but due to the limitations of the modeling techniques, only ∼15% of the human transcripts are endowed with a reliable protein structure prediction.
ASPICdb aims at filling the gap of structural and functional annotation of protein splicing variants, by adopting a set of analysis and prediction tools that do not rely only on annotation transfer by sequence similarity. It provides a thorough computational annotation of predicted human protein variants including PFAM domains (25), N-terminal signal peptides, GPI-anchor propeptides, transmembrane domains, subcellular localization and other features, also reporting the relevant crosslinks to UniprotKB/Swissprot (26) and PDB databases (27). A comprehensive annotation of the domain architecture and other structural features could also be extremely useful to critically assess the reliability of the functional classification provided the GO System (25), which still neglects much of the relevant information for alternative splicing products.
In addition, in consideration of the fragmented nature of the available transcript data, the new version of ASPicDB include the annotation of CAGE tags (28) in order to identify truly transcription initiation sites and discriminate between full-length isoforms using alternative transcription initiations and 5′-partial transcripts for which a full-length CDS and the encoded protein cannot be reliably predicted.
ANNOTATION PIPELINE OF HUMAN PROTEIN VARIANTS
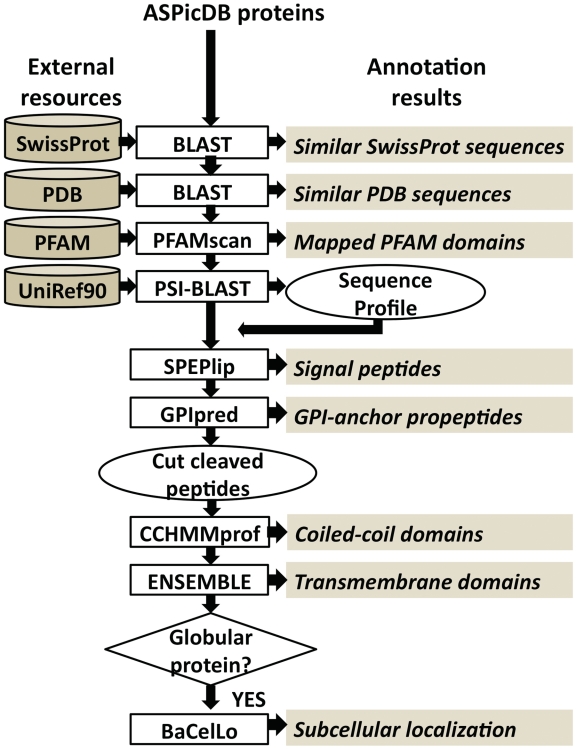
The computational pipeline implemented for supplementing the ASPicDB protein sequences with functional and structural annotations is represented in Figure 1 and integrates several state-of-the-art tools for similarity search and for machine-learning based prediction of protein features starting from residue sequence.
Figure 1.
Pipeline for the annotation of alternative transcripts.
For each one of the 256 939 protein variants coming from 17 191 human genes, a first layer of annotation consists in the retrieval of similar sequences from the two major repositories containing well-characterized proteins, namely: (i) the UniProtKB/SwissProt data base (26) (rel. 2010_07, June 2010), that contains 547 011 protein sequences with curated annotations, including 517 802 principal entries and 29 209 splicing variants (UniProt Consotium, 2010); (ii) the Protein Data Bank (rel. July 2010), that contains resolved three dimensional structures for 50 171 different protein sequences (29).
Similarity searches were performed with BLAST (30) setting the E-value threshold to 10−3.
A second layer of annotation is obtained by mapping the structural and functional domains collected in the PFAM-A database (rel. 24.0, October 2009) that contains curated multiple sequence alignments based on hidden Markov models (HMM) for 8691 families, 2985 domains, 162 repeats and 74 motifs (25). The PFAM models were mapped on the ASPicDB protein sequences by means of the pfam_scan.pl program (ftp://ftp.sanger.ac.uk/pub/databases/Pfam/Tools/), based on HMMER3.0 (31).
The third layer of annotation results from the integration of several predictors based on machine learning tools, such as neural networks, hidden Markov models, support vector machines and conditional random fields. Since most of the methods take advantage of the evolutionary information encoded in sequence profiles, we compiled them starting from the similar sequences retrieved with two PSI-BLAST iterations (setting the E-value threshold to 10−3) from the UniRef90 data set consisting of 6 955 504 sequences (July 2010). The first predicted features are the presence of N-terminal signal peptide and of C-terminal GPI-anchor propeptides, with SPEPlip (32) and PredGPI (33), respectively. Both the methods are among the best available predictors, scoring with accuracy as high as 95% the former and 88% the latter. When present, the signal peptide and the propeptide are cleaved from the protein sequence. The presence of coiled-coil domains is predicted with CCHMM-PROF that is able to locate coiled-coil segments in protein sequences with 80% accuracy (34). α-Helical transmembrane domains are then predicted with ENSEMBLE (35), that discriminates transmembrane from globular proteins with false positive and false negative rates both equal to 3%. The same tool is adopted for predicting the number and the position of transmembrane segments along the sequence, with an accuracy of 90% on the protein base. The subcellular localization of globular proteins is predicted with BaCelLo (36), which discriminates four localizations in animals (secretory pathway, cytoplasm, nucleus and mitochondrion) with 74% accuracy.
ASPicDB CONTENT AND ANNOTATION OF PROTEIN VARIANTS
Table 1 reports some statistics on the data contained in the current version of ASPicDB (version 2.0, August 2010) which refers only to human multi-exon genes annotated in NCBI Entrez Gene (37) with at least one RefSeq transcript (38) and the relevant Unigene cluster (39) collecting all available gene-specific cDNA and EST sequences.
Table 1.
Statistics of the ASPicDB content (v2.0, August 2010)
| ASPicDB v2.0 | |
|---|---|
| Genes | 17 191 |
| Transcripts | 319 092 |
| Proteins | 256 939 |
| Exons | 390 886 |
| Splicing sites | 351 345 |
| U2 | 302 164 |
| U12 | 1712 |
| Splicing events | 233 717 |
The number of splicing sites belonging to the U2 or U12 class and of splicing events is also reported.
In the current version of ASPicDB some more features are available including the annotation of the CAGE tags (28) which define truly transcription initiation sites and a comprehensive protein annotation. A total of 12 789 394 CAGE tags have been mapped thus supporting constitutive or alternative transcription start sites. To each transcript variant a ‘unique identifier’ (16) has been associated in order to make possible the unambiguous comparison with alternative transcripts collected in other databases.
All alternative proteins collected in ASPicDB have been compared with UniprotKB/SwissProt (26) and PDB (29) databases. The results of similarity searches are reported in Table 2. Only 17% of the ASPicDB protein sequences are identical to proteins deposited in UniProtKB/SwissProt database. However, 94% of the sequences share significant similarity with proteins annotated in the same database, prompting the possibility of a reliable annotation transfer. Moreover, 54% of ASPicDB sequences are similar to proteins deposited in the PDB suggesting that their structures can be modeled, at least partially.
Table 2.
Annotation of human variants upon similarity and PFAM searches
| Sequence repository | No of proteinsa | No of genesa |
|---|---|---|
| UniProtKB/SwissProt, % | ||
| E-value < 10−3, % | 239 814 (93) | 17 054 (99) |
| Identical, % | 42 601 (17) | 13 043 (76) |
| PDB | ||
| E-value < 10−3, % | 137 528 (54) | 11 062 (64) |
| Identical, % | 1079 (0.4) | 316 (2) |
| PFAM | ||
| All matches, E-value < 10−5, % | 183 483 (71) | 14 205 (83) |
| Complete matches, E-value < 10−5, % | 46 630 (18) | 5621 (33) |
aThe percentages are computed with respect to 256 939 protein variants and 17 191 genes.
A considerable amount of PFAM models map on the ASPicDB sequences (Table 2). On the overall, 71% of sequences match with at least one model. This result is in agreement with the reported sequence coverage on the human proteome of the current PFAM release, which is equal to 72.5% (25). It is worth noticing that, although all the models map with an E-value < 10−5, only 20% of the matches are complete (that is, involve the whole model). A note of caution is necessary when inferring features from partial matches and the actual extent of the match has to be evaluated for each instance.
Table 3 summarizes the results of the annotation process performed with machine learning based predictors. Two percent of proteins were not predicted since they are shorter than 50 residues, 16% of proteins are predicted as transmembrane and 82% are predicted as globular. Among the globular proteins, 12% are predicted as secreted, 35% as cytoplasmic, 27% as globular and 8% as mitochondrial. Signal peptides and GPI-anchor propeptides are predicted in the 12 and 0.7% of the sequences, respectively. Coiled-coil domains are predicted in 1.3% of the proteins. At the gene level, 30 and 92% of genes encode for transmembrane and globular proteins, respectively. Since the sum exceed 100%, it follows that 22% of the genes encode for both globular and transmembrane variants. The same consideration holds for the other annotations as reported in Table 4. The amount of genes predicted to encode for proteins with different subcellular localization achieves 56%. This is partially explained by the fact that BaCelLo scores with an accuracy equal to 74%, which is the lowest among the methods included in the pipeline. Indeed the discrimination between the ‘cytoplasmic’ and the ‘nuclear’ classes is still a difficult task for all subcellular localization predictors (40). When the two classes are merged together, the BaCelLo accuracy increases up to 91%, but the rate of genes encoding for proteins with different localizations is still as high as 44%, suggesting that localization diversity is inherent in the ASPicDB protein variants. The structure of PFAM annotations is also highly variable: 38% of genes encode for variants matching with different number and/or type of PFAM models. Altogether, results listed in Table 4 suggest that alternative transcripts can encode for proteins endowed with different structural and functional features. ASPicDB provides a unique resource reporting the annotation of alternative splicing variants at the protein level and an interface enabling the discovery of such differences.
Table 3.
Machine learning-based prediction of the human proteins deposited ASPicDB
| Annotation | No. of proteinsa | No. of genesa |
|---|---|---|
| Type | ||
| Globular, % | 210 608 (82) | 15 513 (90) |
| Transmembrane, % | 41 561 (16) | 5439 (32) |
| Localization (globular proteins) | ||
| Secretory pathway, % | 31 917 (12) | 7348 (43) |
| Cytoplasm, % | 90 046 (35) | 10 327 (60) |
| Nucleus, % | 69 167 (27) | 8183 (48) |
| Mitochondrion, % | 19 478 (8) | 4698 (27) |
| Domains | ||
| Signal peptide, % | 30 508 (12) | 5153 (30) |
| GPI-anchor propeptide, % | 1673 (0.7) | 629 (4) |
| Coiled-coil segments, % | 3423 (1.3) | 497 (2.8) |
aThe percentages are computed with respect to 256 939 protein variants and 17 191 genes.
Table 4.
Differences among alternative proteins encoded by the same human gene
| Annotation | No. of genesa, % |
|---|---|
| Type (globular/transmembrane) | 3817 (22) |
| Subcellular localization (globular proteins) | 9593 (56) |
| Presence of signal peptide | 3939 (23) |
| Presence of GPI-anchor propeptide | 591 (3.4) |
| Presence of coiled-coil domains | 464 (2.7) |
| Number of transmembrane helices | 2140 (12) |
| PFAM models (all matches) | 6575 (38) |
aThe percentages are computed with respect to 17 191 genes.
ASPicDB RETRIEVAL INTERFACE
ASPicDB can be accessed though simple or advanced query forms. The simple query form allows the user to obtain the splicing pattern of one or more genes selected according to several criteria (e.g. HGNC name, RefSeq or Unigene accession IDs, etc.). The advanced query form allows the user to search for (i) genes, (ii) transcripts; (iii) exons; (iv) splicing sites; and (v) proteins, fulfilling different criteria (e.g. exons in a given length range, etc.). Depending on the choice separate query forms appear. The ‘gene’, ‘transcript’ and ‘splicing sites’ query forms have been described previously (15) whereas the ‘exon’ and ‘protein’ query forms are novel features of this version of ASPicDB. The exon query form allows the user to select exons in a given length range, belonging to a specific type (initial, internal or teminal), flanked by specific splicing sites or associated to one or more Affimetrix ExonArray probeset IDs.
The ‘protein’ query form allows the retrieval of transcripts encoding proteins isoforms of a specific class (e.g. globular or transmembrane), subcellular localization (e.g. mitochondrion, nucleus, secretory, cytoplasm) or containing one or more features, including occurrence and number of PFAM or transmembrane domains, GPI-anchor propeptides, signal peptides. Finally, it is also possible to retrieve genes encoding for alternative proteins that show differences in the above mentioned features.
ASPicDB OUTPUT
After a simple or advanced query has been submitted the output for each selected gene is shown which is organized in eight panels.
Gene information reports a summary of the genomic and transcript data used by ASPIC to generate the prediction, downloadable by the user and links to other popular prediction programs such as ASAP2 (21), ASD (20) and ACEVIEW (41) as well as to ASPIC results for orthologous genes in other species.
Gene structure view provides a schematic graphical view of the gene structure including all predicted exons/introns.
Predicted transcripts show a graphical representation of the assembled transcripts with predicted annotations of 5′-UTR, CDS and 3′-UTR, CAGE tag mapping, Premature Termination Codons (PTC) and polyA sites.
Transcript table lists the details of all predicted alternative transcripts including their length, number of exons and presence of a protein coding sequence. The ‘variant type’ column lists all the alternative splicing events using a RefSeq mRNA as the reference transcript. The transcript signature is also reported which consists in a unique ID for alternatively spliced variants generated according to (16).
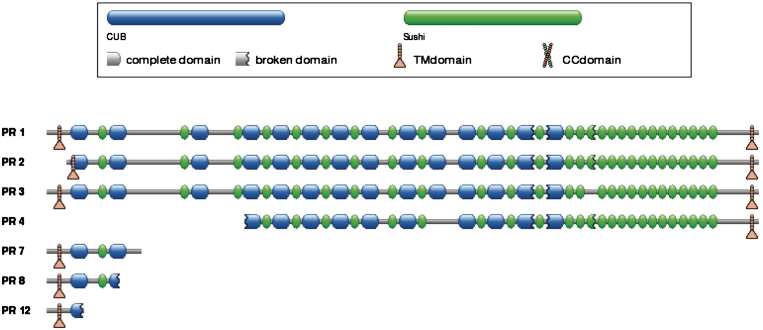
Predicted proteins show a graphical representation of the encoded proteins with matching domains (Figure 2). For each mapped domain the sequence coordinates are reported and different symbols indicate whether the mapping involves the complete domain or only a part of it.
Protein table lists the predicted features of the alternative proteins that include: (i) the best hits obtained from the similarity searches against the UniProtKB/SwissProt and PDB databases, along with the identity value and coverage of the alignment with respect to both the query and the subject sequence lengths; (ii) the features predicted by the pipeline based on machine-learning tools.
Predicted splice sites shows the multiple alignment between the genomic sequence and the expressed sequences (i.e. mRNAs and ESTs) near the boundaries (splice sites) of all predicted introns.
Intron table lists all predicted introns and their relevant features; All results can be also downloaded by the user in textual format following the ‘gene transfer format’ (GTF) (see the Gene Information panel).
Figure 2.
‘Predicted proteins’ panel for gene CSMD3 (CUB and Sushi multiple domains 3). The gene is predicted to encode for 12 transcripts and 7 different protein sequences. Variants labeled as PR1, PR2 and PR3 are identical to the isoforms reported in the CSMD3_HUMAN entry of SwissProt/UniprotKB. Two more variants are reported in that file, although lacking of experimental annotations. Several repetitions of Sushi and CUB domains are predicted with PFAM (25) and represented with symbols indicating whether the model is completely or partially mapped to the sequence. The two transmembrane helices are predicted with ENSEMBLE (35).
After a query at the gene, transcript, exon, protein or splice site level has been completed, the user can also download specific sets of sequences in FASTA format for further analyses, e.g. genes, transcripts, exons, proteins, 5′-UTRs, coding sequences, 3′-UTRs, introns as well as sequence regions surrounding splice site boundaries.
FUTURE PERSPECTIVES
ASPicDB is an ongoing project and we plan to further develop it in the next releases. In particular we plan to add specific annotations on splicing regulatory elements and their interacting RNA-binding proteins located both in exonic and intronic regions. We also plan to update alternative splicing prediction by using the huge amount of RNA-Seq data which are now being produced by next generation sequencing, possibly annotating splicing events as constitutive or tissue-specific. Furthermore, literature-screened splicing patterns related to diseases will be annotated as they represent potential molecular biomarkers and possible targets for therapy. Finally, the inclusion in the database of data related to other organisms will certainly favor a better understanding of the alternative splicing process through comparative analyses.
FUNDING
Ministero dell’Istruzione, dell’Università e della Ricerca: Fondo Italiano Ricerca di Base: ‘Laboratorio Internazionale di Bioinformatica’ (LIBI); Laboratorio di Bioinformatica per la Biodiversità Molecolare (MBLAB) and Telethon (project GGP01658). Funding for open access charge: Ministero dell’Università e della Ricerca: Fondo Italiano Ricerca di Base: ‘Laboratorio Internazionale di Bioinformatica’ (LIBI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 3.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 5.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 6.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 7.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 8.Pettigrew CA, Brown MA. Pre-mRNA splicing aberrations and cancer. Front. Biosci. 2008;13:1090–1105. doi: 10.2741/2747. [DOI] [PubMed] [Google Scholar]
- 9.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J. Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 10.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 11.Boguski MS, Lowe TM, Tolstoshev CM. dbEST–database for “expressed sequence tags”. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 12.Bonizzoni P, Rizzi R, Pesole G. ASPIC: a novel method to predict the exon-intron structure of a gene that is optimally compatible to a set of transcript sequences. BMC Bioinformatics. 2005;6:244. doi: 10.1186/1471-2105-6-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castrignano T, Rizzi R, Talamo IG, De Meo PD, Anselmo A, Bonizzoni P, Pesole G. ASPIC: a web resource for alternative splicing prediction and transcript isoforms characterization. Nucleic Acids Res. 2006;34:W440–W443. doi: 10.1093/nar/gkl324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonizzoni P, Mauri G, Pesole G, Picardi E, Pirola Y, Rizzi R. Detecting alternative gene structures from spliced ESTs: a computational approach. J. Comput. Biol. 2009;16:43–66. doi: 10.1089/cmb.2008.0028. [DOI] [PubMed] [Google Scholar]
- 15.Castrignano T, D’Antonio M, Anselmo A, Carrabino D, D'Onorio De Meo A, D'Erchia AM, Licciulli F, Mangiulli M, Mignone F, Pavesi G, et al. ASPicDB: a database resource for alternative splicing analysis. Bioinformatics. 2008;24:1300–1304. doi: 10.1093/bioinformatics/btn113. [DOI] [PubMed] [Google Scholar]
- 16.Riva A, Pesole G. A unique, consistent identifier for alternatively spliced transcript variants. PLoS ONE. 2009;4:e7631. doi: 10.1371/journal.pone.0007631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–4886. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PL, Albrecht M, Hegyi H, Giorgetti A, et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamm S, Riethoven JJ, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim N, Alekseyenko AV, Roy M, Lee C. The ASAP II database: analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35:D93–D98. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foissac S, Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007;35:W297–W299. doi: 10.1093/nar/gkm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeda J, Suzuki Y, Sakate R, Sato Y, Gojobori T, Imanishi T, Sugano S. H-DBAS: human-transcriptome database for alternative splicing: update 2010. Nucleic Acids Res. 2010;38:D86–D90. doi: 10.1093/nar/gkp984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birzele F, Kuffner R, Meier F, Oefinger F, Potthast C, Zimmer R. ProSAS: a database for analyzing alternative splicing in the context of protein structures. Nucleic Acids Res. 2008;36:D63–D68. doi: 10.1093/nar/gkm793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 27.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. CAGE: cap analysis of gene expression. Nat. Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 29.Dutta S, Burkhardt K, Swaminathan GJ, Kosada T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Methods Mol. Biol. 2008;426:81–101. doi: 10.1007/978-1-60327-058-8_5. [DOI] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 32.Fariselli P, Finocchiaro G, Casadio R. SPEPlip: the detection of signal peptide and lipoprotein cleavage sites. Bioinformatics. 2003;19:2498–2499. doi: 10.1093/bioinformatics/btg360. [DOI] [PubMed] [Google Scholar]
- 33.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartoli L, Fariselli P, Krogh A, Casadio R. CCHMM_PROF: a HMM-based coiled-coil predictor with evolutionary information. Bioinformatics. 2009;25:2757–2763. doi: 10.1093/bioinformatics/btp539. [DOI] [PubMed] [Google Scholar]
- 35.Martelli PL, Fariselli P, Casadio R. An ENSEMBLE machine learning approach for the prediction of all-alpha membrane proteins. Bioinformatics. 2003;19(Suppl. 1):i205–i211. doi: 10.1093/bioinformatics/btg1027. [DOI] [PubMed] [Google Scholar]
- 36.Pierleoni A, Martelli PL, Fariselli P, Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics. 2006;22:e408–e416. doi: 10.1093/bioinformatics/btl222. [DOI] [PubMed] [Google Scholar]
- 37.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuo D, Zhao WD, Wright FA, Yang HY, Wang JP, Sears R, Baer T, Kwon DH, Gordon D, Gibbs S, et al. Assembly, annotation, and integration of UNIGENE clusters into the human genome draft. Genome Res. 2001;11:904–918. doi: 10.1101/gr.164501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casadio R, Martelli PL, Pierleoni A. The prediction of protein subcellular localization from sequence: a shortcut to functional genome annotation. Brief. Funct. Genomic Proteomic. 2008;7:63–73. doi: 10.1093/bfgp/eln003. [DOI] [PubMed] [Google Scholar]
- 41.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1):S12, 1–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]