Abstract
Cell line identification is emerging as an essential method for every cell line user in research community to avoid using misidentified cell lines for experiments and publications. IGRhCellID (http://igrcid.ibms.sinica.edu.tw) is designed to integrate eight cell identification methods including seven methods (STR profile, gender, immunotypes, karyotype, isoenzyme profile, TP53 mutation and mutations of cancer genes) available in various public databases and our method of profiling genome alterations of human cell lines. With data validation of 11 small deleted genes in human cancer cell lines, profiles of genomic alterations further allow users to search for human cell lines with deleted gene to serve as indigenous knock-out cell model (such as SMAD4 in gene view), with amplified gene to be the cell models for testing therapeutic efficacy (such as ERBB2 in gene view) and with overlapped aberrant chromosomal loci for revealing common cancer genes (such as 9p21.3 homozygous deletion with co-deleted CDKN2A, CDKN2B and MTAP in chromosome view). IGRhCellID provides not only available methods for cell identification to help eradicating concerns of using misidentified cells but also designated genetic features of human cell lines for experiments.
INTRODUCTION
Cell lines are important and essential reagents for almost every experiment in biomedical research. Because of indefinite growing capability and extensive use in research community, cell lines are frequently contaminated and misidentified in literatures (1–3). It has been recommended that researchers should provide authenticities of their experimental cell lines before publications (4–6).
The problem of using misidentified cell lines in published literatures has been recognized for several decades. It has been reported by international cell repositories that the incidence of cell line misidentification has been estimated in between 16% and 36% based on analysis of submitted cell lines (4). The most notorious example is the false description of HeLa cervical cancer cell line to several different origins and cell types including Chang liver as ‘normal liver cell’, KB as ‘oral cancer cell’, HEp-2 as ‘laryngeal cancer cell’, Int-407 as ‘non-transformed intestinal epithelial cell’ and so on (7–9). The consequences of using misidentified and contaminated cell lines not only generate erroneous and misleading results but also waste research funding and delay scientific progression. Currently, there is a list of contaminated or misidentified cell lines released in the websites of the international cell repositories to avoid using incorrect cell lines for experiments (5).
For cell identification, several methods were developed to detect cell line contamination or misidentification including isoenzyme analysis, karyotyping, human leukocyte antigen (HLA)-typing, immunotyping, DNA fingerprinting and short tandem repeat (STR) profiling. These methods can distinguish and match the cell line identity to the cell line specific profiles, but with various levels of ambiguity and limitations (4). Among these methods, STR profiling of human cell lines adopted from routing assays of paternity testing and forensic analysis is becoming the most recommended method for cell identification (4,10). A database, Cell Line Integrated Molecular Authentication (CLIMA), collecting the STR profiling of human cell lines is currently available for scientific community (11). However, there are disadvantages of STR profiling in its application for cell identification. First of all, the major limitation for STR profiling is its capability to detect cell contamination from other species because PCR primers of STR markers for authenticity were designed based on human sequence. Second, since majority of cell lines were established from malignant tissues, the gain and loss of cancer genomes increase the ambiguity and reduce the power to match a specific STR profile. Third, STR profiling is unable to distinguish sub-lines derived from the same cell line due to identical STR alleles. Finally, STR profiling requires commercial reagents, expensive instruments and genotyping software for data interpretation. Unless the cost of genotyping can be dramatically reduced and the genotyping experiments can be accessed in nearby core facility, using current methods including STR profiling for routing cell identification might not be able to ease the persistent problem of cell misidentification in our research community.
Since the efforts to eradicate the misidentification problem is unsuccessful, IGRhCellID is established to provide not only STR profiles of human cell lines as the most recommended assay for cell authenticity but also other authentic tools with conventional laboratory PCR or DNA sequencing assays for routing examination of proper cell identification. In addition, IGRhCellID can also allow researchers to search the available cell lines with designated genetic features and to identify common altered loci and genes overlapped in multiple cell lines.
DATABASE CONSTRUCTION
IGRhCellID database contains integrated genomic information of 520 human cell lines annotated with eight different methods for cell identification. The conventional methods including cell line information of STR profile, gender, immunotypes, karyotype and isoenzyme profile were downloaded from common international cell repositories including ATCC, DSMZ, JCRB, ECACC and RIKEN BioResource Center. In addition, we provided cell line information of TP53 mutation data from UMD TP53 mutation database (12,13), somatic mutation data from Catalogue of Somatic Mutations in Cancer (COSMIC) database (14) and genome-wide amplicon and homozygous deletion (HD) profile from our laboratory. We recently established a comprehensive protocol to analyze copy number alterations (CNA) in cancer genomes using high density single nucleotide polymorphism (SNP) arrays with non-paired reference genomes (15). Based on our protocol, we were able to validate known and identify novel amplicons and HDs in 23 cancer cell lines and further identify novel cancer genes in hepatocellular carcinomas.
For annotation of genome-wide amplicons and HDs in human cell lines, we downloaded 520 independent genotyping data of Affymetrix 250 K, 500 K and 6.0 (1800 K) SNP GeneChip array sets of human cell lines from Array Data Management System at National Cancer Institute of USA (caArray, 338 cancer cell lines) (16), Gene Expression Omnibus database (GEO, 182 cell lines) (17) and International HapMap project as normal reference control data. By using dChip analysis software (18), we smoothed each SNP data intensity by three continuous SNPs to create an inferred copy number (ICN) for each SNP and defined amplicons and HDs in greater than 3 and less than 0.5 ICN, respectively. Together, we identified a total of 13 840 927 amplified SNPs and 182 343 loss SNPs located in amplicons and HDs, respectively in 520 human cell lines. To check our analysis protocol and data quality, we examined 11 known genes (LRP1B, FHIT, PARK2, PTPRD, CDKN2B, CDKN2A, PTEN, WWOX, CREBBP, TP53 and SMAD4) with reported small deletions in published literatures for validation (19–29). In 49 previously reported deletion events of above 11 genes in cancer cell lines, we have 96% (47/49) validation rate (Supplementary Table S1). Furthermore, we downloaded and integrated 950 microarray gene expression data sets of the 358 cell lines from caArray to show the concordance of altered gene expression with the corresponding aberrant amplicons and HD regions.
RESULTS
Cell identification
Cell lines can be selected by quick search, by searching the name in alphabetic order or by searching the categories of cell line origin based on body location/systems in National Cancer Institute. After selecting a cell line in cell line view, in addition to common STR profiling and other conventional methods, users can search its genome-wide altered SNPs in amplicons and HDs along the physical and cytogenetic locations of chromosomes (Figure 1). In genomic alteration profile of a selected cell line, user can continuously click on the alteration (green lines strand for HD and red lines strand for amplification) and retrieve SNPs for cell identification with multiple PCR or quantitative PCR reactions. Furthermore, the existence of somatic mutations in TP53 (UMD TP53 database) or other cancer genes (COSMIC) can provide additional support for authenticity.
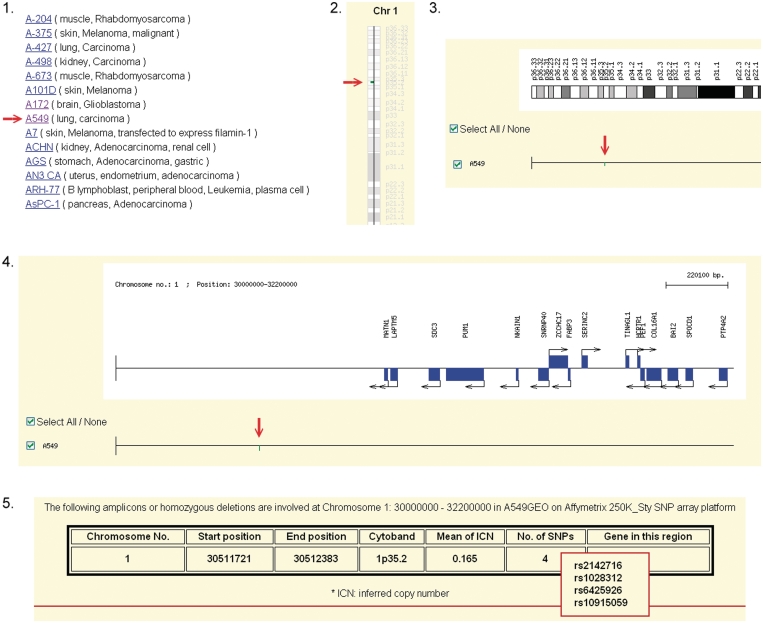
Figure 1.
A tutorial example to find homozygous deleted SNPs in a cell line. In step 1, users can select a cell line by the name in quick search, in alphabetic order or in categories based on NCI cancers by body location/systems. By clicking A549 as an example, in step 2, users will retrieve the profile of genome-wide amplicons and HDs (chromosome 1 as an example) along with the available authentic tools for A549. In step 3, by clicking the HD on chromosome 1p35.2 as an example, the detail localizations of HD and genes are presented. In step 4, users can further click on the HD to display the raw data. In step 5, the genes and SNPs located in HD are presented. By moving curser to the ‘No. of SNP’ or ‘gene in this region’, user can observe the SNPs and genes residing in this HD, respectively. These SNPs will allow users to retrieve primers information in NCBI dbSNP for authentic experimental designs.
Altered genes and loci in cell lines
The profiles of genome-wide amplicons and HDs not only provide a new authentic method of a cell line but also allow for selection of a designated cell line with requested genetic features (in gene or intergenic view). Moreover, common somatic alterations shared in multiple cell lines across cancer types can be displayed in chromosome view. For instance, after selection of cell lines in gene view, users can search a gene and align its genetic alterations in one or multiple cell lines. By using quick examples provided in IGRhCellID, users can find that SMAD4 a tumor suppressor gene in pancreatic carcinoma is deleted in FaDu (a pharynx squamous carcinoma cell line), COLO 201 (colorectal adenocarcinoma cell line) and SW 1573 (lung alveolar carcinoma cell line) with concordance of down regulation in gene expression. Only SMAD4 HD in FaDu was reported before but not COLO 201 and SW 1573 cells (19). The integrated information will provide the unique SMAD4 HD cell lines as human indigenous knock-out model for biological studies.
In intergenic view, after selection of cell lines, users can zoom out the aberrant amplicon or HD region to examine the boundary of aberrant locus and its affected neighboring genes. For instance, ERBB2 also called HER2 or NEU is a known oncogene in human breast cancer. When search ERBB2 in 26 breast cancer cell lines, users can observe only 20% (6/26) cell lines with amplification in the region. After selecting six amplified cell lines and zoom out to a 1 Mb region surrounding ERBB2, users will observe boundaries in some breast cell lines including a smallest amplicon in HCC2218 cells containing eight genes and the largest amplicon in UACC-812 cells containing 29 genes. The amplification of oncogenic ERBB2 in these cell lines will provide not only positive control cell lines for ERBB2 expression but also natural cell models for studying the therapeutic efficacy of drugs against ERBB2 over-expression in breast cancer cells.
In chromosome view, IGRhCellID also allows users to search a cytogenetic region with overlapped somatic alterations in selected human cell lines across cancer types in detail resolution to gene and SNP levels. As indicated in our tutorial example, aberrant chromosome nine in six cell lines classified in endocrine system (five thyroid carcinoma and one adrenal gland carcinoma cell lines) were selected and aligned in chromosome view. Apparently, one overlapped HD region on 9p21.3 was detected in three cell lines. After clicking on the chromosome to enlarge the view, three known genes CDKN2A, CDKN2B and MTAP were co-deleted in the HD region.
DISCUSSION
In addition to providing information of STR profiles of 520 human cell lines, IGRhCellID integrated other authentic tools and genomic alterations for helping researchers to validate their experimental cell lines using conventional methods, to select suitable cell lines with proper genetic background for their experimental designs and to support the long standing efforts for eradicating the concerns of using misidentified cell lines. Using authentic tools to maintain correct cell identification is even more critical in fields of establishing and using stem cell lines for therapeutic applications. Although integration of available authentic tools and profiles of genomic alterations of human cell lines in IGRhCellID provides convenient and accessible methods for cell identification, additional efforts to apply statistic analysis for obtaining the discrimination power of cell authenticity using either one or combined authentic methods should be studied. Nevertheless, IGRhCellID will continue to collect available authentic data for all available cell lines derived from human and other species. Our efforts should help resolving the crisis of using misidentification cell lines and improve the selection of proper cell lines for designated experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Program for Genomic Medicine (NRPGM), National Science Council, Taiwan (grant numbers NSC98-3112-B-001-004 and NSC98-3112-B-001-031). Funding for open access charge: Institute of Biomedical Sciences, Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Phuchareon J, Ohta Y, Woo JM, Eisele DW, Tetsu O. Genetic profiling reveals cross-contamination and misidentification of 6 adenoid cystic carcinoma cell lines: ACC2, ACC3, ACCM, ACCNS, ACCS and CAC2. PLoS One. 2009;4:e6040. doi: 10.1371/journal.pone.0006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiong E, Dadbin A, Harris LD, Sabichi AL, Grossman HB. The use of short tandem repeat profiling to characterize human bladder cancer cell lines. J. Urol. 2009;181:2737–2748. doi: 10.1016/j.juro.2009.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenzi PL, Reinhold WC, Varma S, Hutchinson AA, Pommier Y, Chanock SJ, Weinstein JN. DNA fingerprinting of the NCI-60 cell line panel. Mol. Cancer Ther. 2009;8:713–724. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Cell Line misidentification: the beginning of the end. Nat. Rev. Cancer. 2010;10:441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 5.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 6.Katsnelson A. Biologists tackle cells' identity crisis. Nature. 2010;465:537. doi: 10.1038/465537a. [DOI] [PubMed] [Google Scholar]
- 7.Lucey BP, Nelson-Rees WA, Hutchins GM. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch. Pathol. Lab. Med. 2009;133:1463–1467. doi: 10.5858/133.9.1463. [DOI] [PubMed] [Google Scholar]
- 8.Ogura H, Fujii R, Hamano M, Kuzuya M, Nakajima H, Ohata R, Mori T. Detection of HeLa cell contamination–presence of human papillomavirus 18 DNA as HeLa marker in JTC-3, OG and OE cell lines. Jpn. J. Med. Sci. Biol. 1997;50:161–167. doi: 10.7883/yoken1952.50.161. [DOI] [PubMed] [Google Scholar]
- 9.Ogura H, Yoshinouchi M, Kudo T, Imura M, Fujiwara T, Yabe Y. Human papillomavirus type 18 DNA in so-called HEP-2, KB and FL cells–further evidence that these cells are HeLa cell derivatives. Cell Mol. Biol. 1993;39:463–467. [PubMed] [Google Scholar]
- 10.Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, Furtado M, Kline MC, Kohara A, Los GV, et al. Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues. In Vitro Cell Dev. Biol. Anim. 2010 doi: 10.1007/s11626-010-9333-z. doi:10.1007/s11626-010-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res. 2009;37:D925–D932. doi: 10.1093/nar/gkn730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beroud C, Soussi T. The UMD-p53 database: new mutations and analysis tools. Hum. Mutat. 2003;21:176–181. doi: 10.1002/humu.10187. [DOI] [PubMed] [Google Scholar]
- 13.Cariello NF, Beroud C, Soussi T. Database and software for the analysis of mutations at the human p53 gene. Nucleic Acids Res. 1994;22:3549–3550. [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CF, Hsu EC, Lin KT, Tu PH, Chang HW, Lin CH, Chen YJ, Gu DL, Lin CH, Wu JY, et al. Overlapping high resolution copy number alterations in cancer genomes identified putative cancer genes in hepatocellular carcinoma. Hepatology. 2010;52:1690–1701. doi: 10.1002/hep.23847. [DOI] [PubMed] [Google Scholar]
- 16.McCusker JP, Phillips JA, Gonzalez Beltran A, Finkelstein A, Krauthammer M. Semantic web data warehousing for caGrid. BMC Bioinformatics. 2009;10(Suppl. 10):S2. doi: 10.1186/1471-2105-10-S10-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohno T, Otsuka A, Girard L, Sato M, Iwakawa R, Ogiwara H, Sanchez-Cespedes M, Minna JD, Yokota J. A catalog of genes homozygously deleted in human lung cancer and the candidacy of PTPRD as a tumor suppressor gene. Genes Chromosomes Cancer. 2010;49:342–352. doi: 10.1002/gcc.20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solomon DA, Kim JS, Ressom HW, Sibenaller Z, Ryken T, Jean W, Bigner D, Yan H, Waldman T. Sample type bias in the analysis of cancer genomes. Cancer Res. 2009;69:5630–5633. doi: 10.1158/0008-5472.CAN-09-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishimoto M, Kohno T, Okudela K, Otsuka A, Sasaki H, Tanabe C, Sakiyama T, Hirama C, Kitabayashi I, Minna JD, et al. Mutations and deletions of the CBP gene in human lung cancer. Clin. Cancer Res. 2005;11:512–519. [PubMed] [Google Scholar]
- 22.Wang F, Denison S, Lai JP, Philips LA, Montoya D, Kock N, Schule B, Klein C, Shridhar V, Roberts LR, et al. Parkin gene alterations in hepatocellular carcinoma. Genes Chromosomes Cancer. 2004;40:85–96. doi: 10.1002/gcc.20020. [DOI] [PubMed] [Google Scholar]
- 23.Pineau P, Marchio A, Cordina E, Tiollais P, Dejean A. Homozygous deletions scanning in tumor cell lines detects previously unsuspected loci. Int. J. Cancer. 2003;106:216–223. doi: 10.1002/ijc.11214. [DOI] [PubMed] [Google Scholar]
- 24.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JE. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc. Natl Acad. Sci. USA. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang SI, Parsons R, Ittmann M. Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin. Cancer Res. 1998;4:811–815. [PubMed] [Google Scholar]
- 26.Druck T, Hadaczek P, Fu TB, Ohta M, Siprashvili Z, Baffa R, Negrini M, Kastury K, Veronese ML, Rosen D, et al. Structure and expression of the human FHIT gene in normal and tumor cells. Cancer Res. 1997;57:504–512. [PubMed] [Google Scholar]
- 27.Takahashi T, Carbone D, Nau MM, Hida T, Linnoila I, Ueda R, Minna JD. Wild-type but not mutant p53 suppresses the growth of human lung cancer cells bearing multiple genetic lesions. Cancer Res. 1992;52:2340–2343. [PubMed] [Google Scholar]
- 28.Wolf D, Rotter V. Major deletions in the gene encoding the p53 tumor antigen cause lack of p53 expression in HL-60 cells. Proc. Natl Acad. Sci. USA. 1985;82:790–794. doi: 10.1073/pnas.82.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matozaki T, Sakamoto C, Matsuda K, Suzuki T, Konda Y, Nakano O, Wada K, Uchida T, Nishisaki H, Nagao M, et al. Missense mutations and a deletion of the p53 gene in human gastric cancer. Biochem. Biophys. Res. Commun. 1992;182:215–223. doi: 10.1016/s0006-291x(05)80133-0. [DOI] [PubMed] [Google Scholar]