Abstract
ZFIN, the Zebrafish Model Organism Database, http://zfin.org, serves as the central repository and web-based resource for zebrafish genetic, genomic, phenotypic and developmental data. ZFIN manually curates comprehensive data for zebrafish genes, phenotypes, genotypes, gene expression, antibodies, anatomical structures and publications. A wide-ranging collection of web-based search forms and tools facilitates access to integrated views of these data promoting analysis and scientific discovery. Data represented in ZFIN are derived from three primary sources: curation of zebrafish publications, individual research laboratories and collaborations with bioinformatics organizations. Data formats include text, images and graphical representations. ZFIN is a dynamic resource with data added daily as part of our ongoing curation process. Software updates are frequent. Here, we describe recent additions to ZFIN including (i) enhanced access to images, (ii) genomic features, (iii) genome browser, (iv) transcripts, (v) antibodies and (vi) a community wiki for protocols and antibodies.
INTRODUCTION
ZFIN is a curated resource for zebrafish biology comprised of the following primary data types: genes, phenotypes, genotypes, gene expression, functional and phenotypic annotations, anatomical structures, orthology, nucleotide and protein sequence associations and reagents such as morpholinos and antibodies. Table 1 lists ZFIN data contents as of July 2010. A tabular presentation of ZFIN’s growth over the years can be accessed from the database (http://zfin.org/zf_info/zfin_stats.html). ZFIN data can be accessed using any of the data-type specific search forms, site search, BLAST, or GBrowse. A comprehensive suite of download files provide a means of accessing large quantities of data for further analysis. Special requests for data reports can be requested from zfinadmn@zfin.org.
Table 1.
Summary of ZFIN data content (July 2010)
| ZFIN data statistics | July 2010 |
|---|---|
| Genes | 30 783 |
| Genes on assembly | 15 341 |
| Transcripts | 25 916 |
| EST/cDNAs | 34 865 |
| Full length cDNA clones (ZGC) | 17 191 |
| Genomic features | 6603 |
| Transgenic features | 2002 |
| Transgenic constructs | 591 |
| Genotypes | 9565 |
| Genes with GO annotations | 15 502 |
| Genes with IEA GO annotations | 11 741 |
| Genes with non-IEA GO annotations | 8728 |
| Total GO annotations | 1 08 694 |
| Morpholinos | 3571 |
| Antibodies | 542 |
| Gene expression patterns | 49 811 |
| Images (phenotypes, expression patterns) | 76 911 |
| Anatomical structures | 2669 |
| Genes with human orthology | 8937 |
| Genes with mouse orthology | 5506 |
| Publications | 13 063 |
| Researchers | 5177 |
| Laboratories | 640 |
| Companies | 125 |
ZFIN participates in regularly scheduled data exchanges, ranging from daily to monthly, with major bioinformatics organizations, such as the Welcome Trust Sanger Institute, Ensembl, NCBI and UniProt resulting in reciprocal links that provide valuable cross-site data integration. These exchanges enhance data accuracy and consistency because curators work continuously to resolve identified discrepancies. In addition, we provide links to many community resources on our home page.
ZFIN’s curation process utilizes bioinformatics community-supported best practices to ensure data are described accurately and consistently. One such practice is the use of standardized nomenclature. ZFIN, in conjunction with the Zebrafish Nomenclature Committee, serves as the authoritative source of gene and allele nomenclature. Standardized nomenclature is essential to unambiguous communication. Zebrafish nomenclature guidelines are coordinated with guidelines used for human and mouse genes. Similarly, standardization of functional and phenotypic gene annotations promotes robust searching and comparisons within and among species. ZFIN’s annotations are based on the structured vocabularies and relationships defined by biological ontologies. These ontologies are evolving resources that require community input to ensure completeness and accuracy. ZFIN collaborates with the bioinformatics community on the development of several ontologies including Gene Ontology [GO; (1)], Cell Ontology [CL; (2)] and Phenotype Quality Ontology [PATO; (3)]. ZFIN also develops and maintains the Zebrafish Anatomical Ontology [ZFA; (4)]. ZFIN is the authoritative source for zebrafish GO annotations. Standardized evidence codes are used to support GO and orthology annotations. All data are attributed to their original sources.
ZFIN encourages comments and suggestions from the community. A ‘Your Input Welcome’ button is provided on every ZFIN data page to facilitate communication. ZFIN curators address incoming questions and data submissions. Requests for new features and enhancements, combined with annual user surveys results, play a key role in determining future directions.
NEW TO ZFIN
Enhanced access to images
ZFIN maintains an extensive repository of annotated figures derived from current literature and data submitted directly to ZFIN by researchers. Recent enhancements, based mainly on user requests, provide increased access to these images and have quickly become ZFIN’s most popular feature.
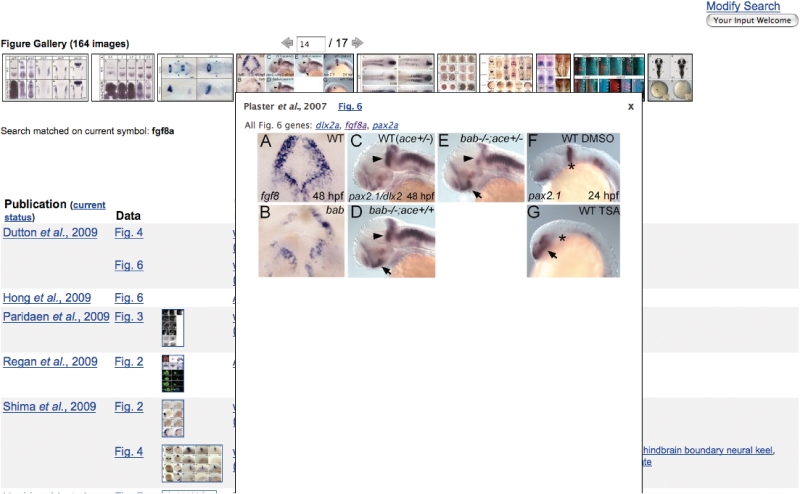
Annotated figures of gene expression patterns are included in this repository. Annotations associate genes, fish, developmental stages and terms from the ZFA ontology to each figure. It is often desirable to browse these figures, using the gene expression search form, for a marker with a particular gene expression pattern. A search for ‘integument’ returns nearly 700 markers. Individually reviewing this large number of matching markers can be a daunting task. A figure gallery thumbnail strip (Figure 1), displaying each figure that matches the search criteria, has been added at the top of each gene expression search results page to provide a quick means to find the desired pattern. Mousing over a thumbnail pops up a larger image with links to detailed information. Controls located above the strip provide navigation through multiple thumbnail strips.
Figure 1.
Gene expression results page depicting the figure galley thumbnail strip with an enlarged thumbnail image showing Figure 6 from Plaster et al., 2007 (9). Thumbnail strip navigation controls have been used to advance to strip 14 of 17.
Terms from the ZFA ontology play an integral role in annotations of phenotype and gene expression data in ZFIN. In addition, definitions and relationships assist in the identification of anatomical structures throughout the development cycle. Each ZFA term is represented on an anatomy page that details synonyms, a definition, developmental stages during which the structure is present, parent and child structures and links to expression and phenotype data for the structure. Images of a select group of approximately 300 anatomical structures are now also available on these pages. Initial images are from Wolfgang Driever’s developmental atlas (http://zfin.org/zf_info/anatomy.html) of the embryo spanning stages prim-5 to Day 5 and images of high quality, labeled mRNA in-situ hybridization studies (5). Plans are underway to add postembryonic images (6) and ZFIN curators will add images from publications as part of our ongoing curation process. Submissions from researchers are also welcome. The anatomy detail page can be accessed using the anatomy browser available from our home page. Extensive data integration produces links to anatomy pages from figure, gene, feature and genotype pages.
Genomic feature page
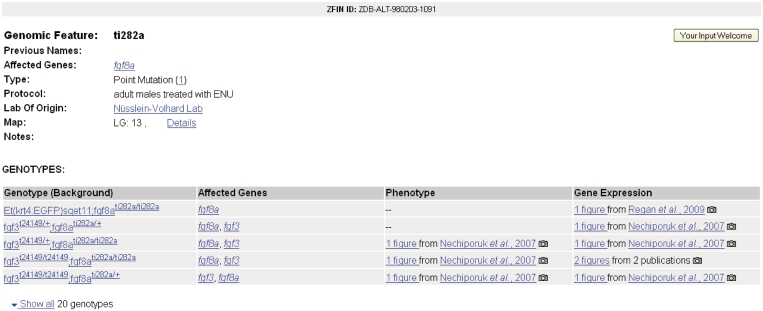
Researchers gain an understanding of how genes function by investigating mutations, or genomic features, of a particular locus. These genomic features include point mutations, insertions and other chromosomal aberrations like deficiencies and translocations. Understanding how a genomic feature affects gene function is key to elucidating biological pathways, disease, gene product interaction and gene regulation. To help researchers begin understanding genomic alterations, the Genomic Feature page (Figure 2) was developed as a resource page that details information about individual genomic features. This page provides information about affected genes, mutation type, protocol used to induce the mutation, lab of origin, mapping details and a table of all associated genotypes with links to phenotype and gene expression data. A camera icon indicates that genotype images are available on ZFIN. Links provide easy navigation to affected gene and genotype pages. The Feature page can also be accessed from links on the Mutant/Morphants/Transgenics Search Results page.
Figure 2.
Screenshot depicting the ti282a feature page. Links are provided to all ti282a data in ZFIN.
GBrowse genome browser
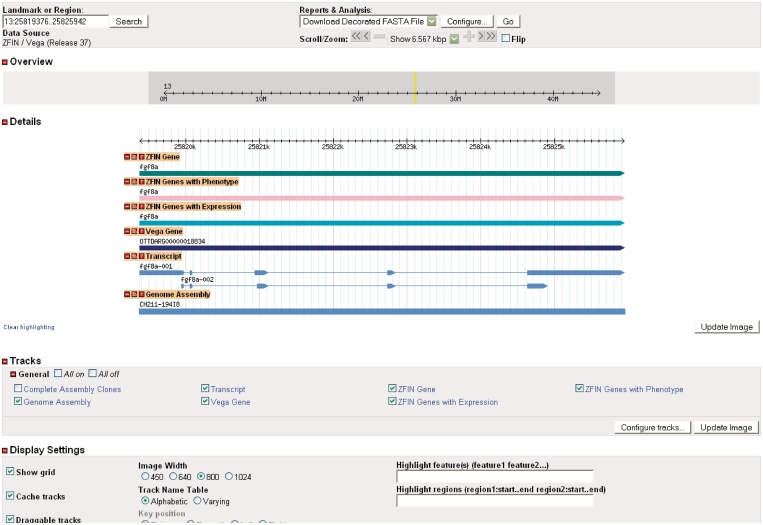
ZFIN has implemented a genome browser, GBrowse, developed by the Generic Model Organism Database (GMOD; http://gmod.org). GBrowse provides a graphical, interactive and customizable web interface to explore the zebrafish genome, including information about genome annotations and the positions of genes on the chromosomes. Tracks are aligned to the current version of the Wellcome Trust Sanger Institute Vertebrate Genome Annotation [Vega; (7)] genome assembly and link to data-specific pages at ZFIN or Vega. Individual tracks show ZFIN genes with phenotype, ZFIN genes with gene expression data, Vega genes, transcripts, BACs used as assembly components, morpholinos and antibodies. GBrowse can be launched from ZFIN’s home page or from the GBrowse interactive graphic found on the gene, transcript and clone detail pages.
Transcript page
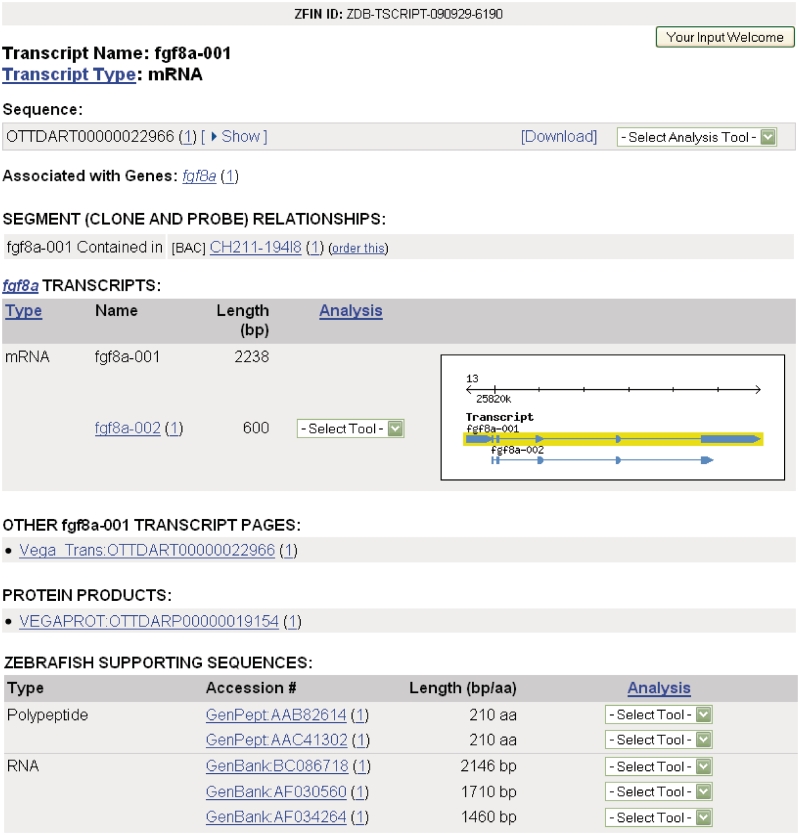
Transcript pages at ZFIN were created to integrate data from the zebrafish whole genome sequence and annotation project at the Welcome Trust Sanger Institute and present users with the set of known transcripts for a gene. This is a necessary first step towards associating expression, function and phenotype data with specific transcripts. The transcript page (Figure 3) currently provides information about the transcript including name, type, associated genes and sequences and other transcripts produced by the gene. Supporting sequences, used as evidence for the transcript by the Sanger annotation team, are also provided. Transcript types are defined using commonly accepted terms from the Sequence Ontology [SO; (8)]. Links to Vega transcript and protein pages are also provided. Sequence searches can be performed at ZFIN, Vega, Ensembl, NCBI and UCSC by selecting the appropriate option from the sequence analysis tools menu. Additionally, the transcript page includes a GBrowse graphic depicting the relevant portion of the genome. Clicking the graphic launches GBrowse that provides access to data for transcripts at the genome level (Figure 4). All transcripts are searchable using the Genes/Markers/Clones search form. Transcript records can also be accessed from gene pages and by sequence similarity search using the BLAST tool at ZFIN.
Figure 3.
The transcript page summarizes transcript-specific data for fgf8a-001.
Figure 4.
The GBrowse view of the genome, centered on fgf8a-001 transcript.
Antibodies
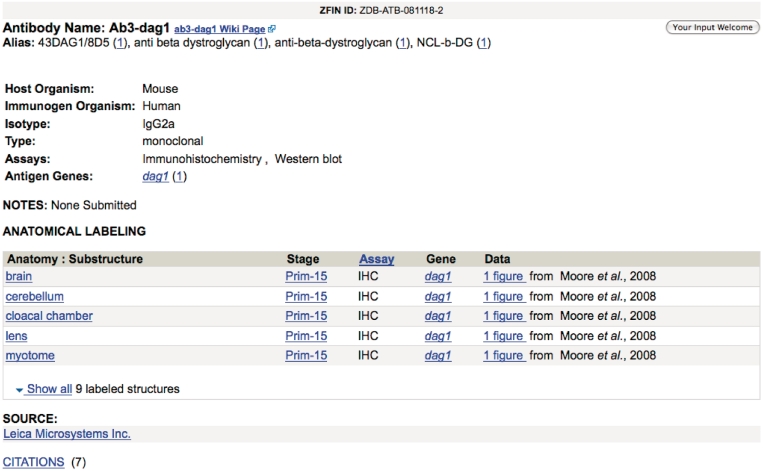
Antibodies are widely used as probes for gene expression studies and as labels of anatomical structures, creating a need for a centralized resource that provides information about antibodies known to work in zebrafish. ZFIN now captures antibody data as part of our literature curation process. An antibody nomenclature convention has been adopted using gene symbols or gene family stem symbols when possible. Antibody data can be retrieved using the new antibody search form. The search form provides options to find antibodies that label specific anatomical structures or that target products of a specific gene. Antibody specific information, such as name, type, host organism and assay may also be used to refine searches. A search results page provides links to matching antibody pages. The information provided on the antibody page (Figure 5) includes the name, aliases, host organism, isotype, type and assays. A summary of wild-type zebrafish labeling patterns includes labeled anatomical structures, developmental stage, assay, associated gene and links to published figures. Sources and usage notes are provided when available. A link is provided to the corresponding community wiki antibody page for community-contributed comments. Links to antibodies that label a specific structure are also available on ZFIN anatomy pages.
Figure 5.
ZFIN Antibody page for Ab3-dag1 antibody.
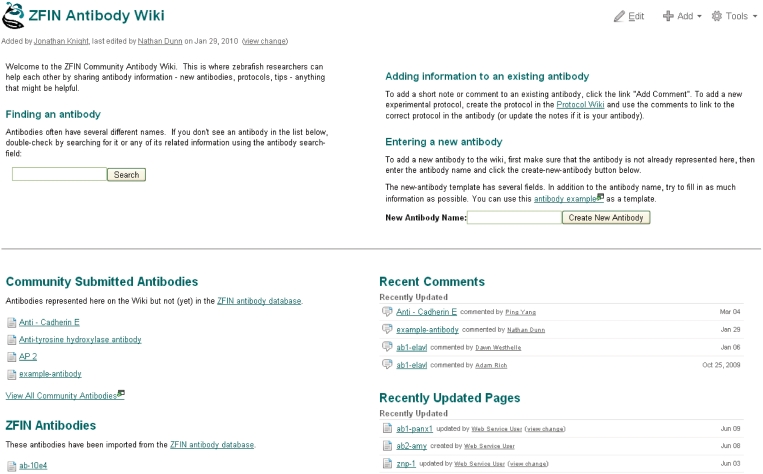
Community wiki
The zebrafish community has an untapped wealth of valuable unpublished knowledge about protocols and reagents. Recently, ZFIN implemented a Community Wiki (http://wiki.zfin.org) for experimental protocols and antibodies to foster community collaboration and sharing in a centralized location. Community provided information supplements, but is not directly integrated with, the published data curated at ZFIN. Contributions provide useful practical tips such as RNAi hairpin design for zebrafish transgenics or preferred fixatives for antibody staining. The wiki format supports easy searching, record creation and editing. A table of contents, the most recent comments and a catalog of the most recently updated pages are also displayed. Login is not required to view information in the wiki. A login link allows registered users to login and contribute new records or comments. The adjacent sign up link can be used by new users to create an account. Instructions for adding new records and comments are provided on the home page for each wiki section (Figure 6). All wiki records can be viewed online or exported as PDF or Word documents.
Figure 6.
Community Wiki Antibody section homepage.
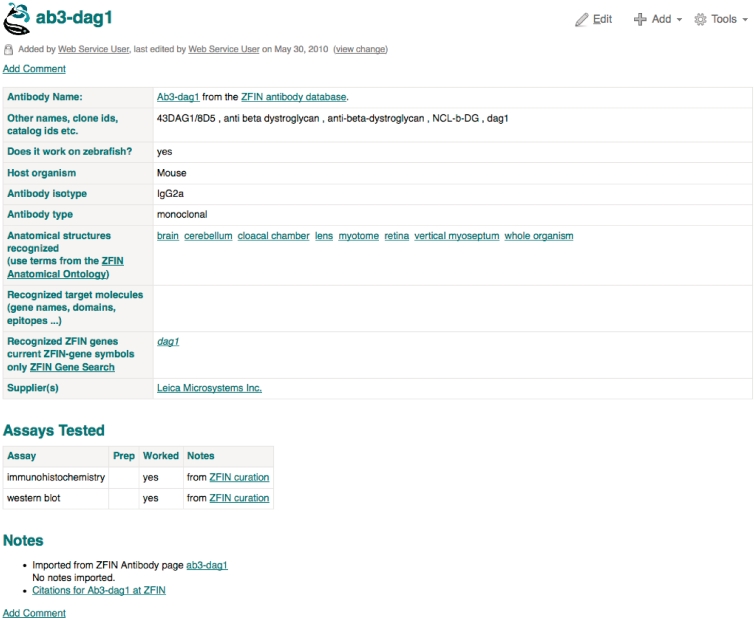
The Protocol Wiki contains protocols from The Zebrafish Book (5th edn), along with protocols shared by researchers through direct submission. Only the submitter can modify a protocol. Other registered users are encouraged to use the comments field to provide additional tips. The Antibody Wiki contains community submitted information, all ZFIN curated antibody records and all antibodies available at the Zebrafish International Resource Center (ZIRC). Individual antibody pages (Figure 7) provide information about antibody names, aliases, catalog IDs, antibody details, structures the antibody labels, target molecules, recognized genes with links to the ZFIN gene page, suppliers, assays tested, notes, comments and links to corresponding ZFIN antibody pages. The community wiki can also be accessed from links provided on ZFIN’s home page or by using the Site Search function provided on ZFIN’s home page.
Figure 7.
Antibody Wiki page for the Ab3-dag1 antibody.
SUBMITTING DATA
ZFIN encourages researchers to share unpublished data using the Phenote software package. Phenote facilitates the annotation of gene expression patterns and mutant phenotypes with the same zebrafish ZFA, GO and PATO ontology terms used by ZFIN curators to annotate published data. The use of common terms provides easy integration into and searching of ZFIN. Phenote is available at http://wwww.phenote.org/download.shtml. All submitted data are directly attributed to their sources.
FUTURE DIRECTIONS
ZFIN will continue detailed curation of current data types. Support will be expanded to include associations between human diseases and zebrafish genes and phenotypes. In addition, ZFIN is designing a new collection of search and browsing tools that will provide enhanced access to the rapidly expanding collection of data. We will soon integrate Intermine (http://www.intermine.org) into the ZFIN site.
CITING ZFIN
Please cite this article for a general reference to the ZFIN database. In addition, the following format is suggested for citing a specific entry in ZFIN. [Type of] data for this paper were retrieved from the Zebrafish Model Organism Database (ZFIN), University of Oregon, Eugene, OR 97403-5274; http://zfin.org/; [the date you retrieved the data cited].
IMPLEMENTATION
ZFIN is currently implemented with IBM/Informix relational database management software. Web-based HTML forms combined with Java/JSP, GWT, JavaScript, Perl and CGI scripts provide access to the database. The community wiki is powered by Atlassian Confluence software (http://www.altassian.com/software/confluence/).
FUNDING
National Institutes of Health (P41 HG002659, R01 HG004838 and R01 HG004834). Funding for open access charge: National Institutes of Health (HG002659).
Conflict of interest statement. None declared.
REFERENCES
- 1.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinement. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thisse B, Pflumio S, Furthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, et al. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402) 2001 ZFIN Direct Data Submission. [Google Scholar]
- 6.Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dyn. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eilbeck K, Lewis S, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plaster N, Sonntag C, Schilling TF, Hammerschmidt M. REREa/Atrophin-2 interacts with histone deacetylase and Fgf8 signaling to regulate multiple processes of zebrafish development. Dev. Dyn. 2007;236:1891–1904. doi: 10.1002/dvdy.21196. [DOI] [PubMed] [Google Scholar]