Abstract
The RIKEN integrated database of mammals (http://scinets.org/db/mammal) is the official undertaking to integrate its mammalian databases produced from multiple large-scale programs that have been promoted by the institute. The database integrates not only RIKEN’s original databases, such as FANTOM, the ENU mutagenesis program, the RIKEN Cerebellar Development Transcriptome Database and the Bioresource Database, but also imported data from public databases, such as Ensembl, MGI and biomedical ontologies. Our integrated database has been implemented on the infrastructure of publication medium for databases, termed SciNetS/SciNeS, or the Scientists’ Networking System, where the data and metadata are structured as a semantic web and are downloadable in various standardized formats. The top-level ontology-based implementation of mammal-related data directly integrates the representative knowledge and individual data records in existing databases to ensure advanced cross-database searches and reduced unevenness of the data management operations. Through the development of this database, we propose a novel methodology for the development of standardized comprehensive management of heterogeneous data sets in multiple databases to improve the sustainability, accessibility, utility and publicity of the data of biomedical information.
INTRODUCTION
Securing the sustainability of databases is one of the most important issues for research institutes, funding agencies and research communities, because the accumulated cost of maintenance becomes a serious burden on the responsible institutes and communities (1). Moreover, the development of technology for biomedical analyses has brought about a dramatic increase in the amount and variety of data and information. The outdating of isolated data is also a serious problem. The association with public data records broadly used in the research community is crucially important to improve the usability and accessibility of data. If data are isolated in the application software without updates from external data, then the data will become increasingly difficult to retrieve by external retrieval systems and will become useless, unnecessarily occupying the storage resource. On the contrary, the integration of a datum with external data will generally increase its usability and value, often promoting unexpected uses and knowledge discovery. In the community of mammalian research, authoritative data are provided by the Mouse Genome Informatics Database (MGI), HUGO Gene Nomenclature Committee (HGNC) and Rat Genome Database (RGD) with nomenclature activities for genes, alleles and strains for each species (2–4). Data from the National Center for Biotechnology Information (NCBI) and Ensembl are also broadly used across species (5,6). The Open Biomedical Ontology (OBO) Consortium, an umbrella body for the developers of life-science ontologies, also provides ontologies developed with the aim of comprehensive annotation of biological information (7,8).
In the mouse genetical research community, these issues have been discussed by international consortia. The Mouse Phenotype Database Integration Consortium (InterPhenome) (http://www.interphenome.org/) and the Coordination and Sustainability of International Mouse Informatics Resources (CASIMIR) (http://www.casimir.org.uk/) have discussed broad issues regarding the integration, coordination, interoperability and sustainability of databases, such as methodologies to integrate phenotype information, the association of phenotype with human disease, models for long-term and financial sustainability for databases and legal issues of data accessibility (9–11). A complete solution to satisfy these multiple and broad requirements at once is desired to ensure the sustainability of databases.
One effective way to reduce the management cost of databases is to share common fundamental infrastructures such as the hardware and application software used in their implementations. Recently, such common operations have been effectively implemented through ‘cloud computing’, which is a type of internet-based computing whereby shared resources, software and information are provided on demand. Cloud computing is often economically beneficial for the facility in terms of the running costs of space, electricity, cooling and staff support (12). If data are properly and continuously managed and integrated with the public data records that are regarded as the de facto standard in the biomedical community, then a common infrastructure could be one of the best ways to achieve cost effectiveness and advanced usability. On the other hand, the ‘semantic web’ offers a series of methods and technologies to develop extensions of the current World Wide Web (WWW) in which information is given well-defined meanings and integrated (13). These technologies include the Resource Description Framework (RDF), a variety of data interchange formats (e.g. RDF/XML, N3, Turtle and N-Triples), and notations such as the RDF Schema (RDFS) and the Web Ontology Language (OWL), all of which are intended to provide a formal description of concepts, terms and relationships within a given knowledge domain. The semantic web is regarded as an integrator across different content and information applications and systems and provides mechanisms for the realization of a common information system. It is also useful for the dissemination of data, providing a standardized framework to describe metadata recommended by the WWW consortium that aids the automated (and also manual) processing of disseminated data to derive meaning from the data. The dissemination of data with standardized metadata risks the extinction of the data and creates the opportunity to promote the discovery of new knowledge. Consequently, the semantic web seems to be suitable as a fundamental technology to implement the common infrastructure.
In this study, we developed a new database, the RIKEN integrated database of mammals, as an official undertaking in RIKEN to integrate heterogeneous mammal-related data in multiple individual databases. This database was constructed on the Scientists’ Networking System (SciNetS: http://www.riken.jp/engn/r-world/info/release/press/2009/090331_2/), a general fundamental system that applies the semantic web technology to provide massive data management, supported by Japan’s national database integration project. In this system, we achieved the top-level ontology-based re-organization of imported data to integrate the typical and instructive knowledge with individual data records. The RIKEN integrated database of mammals is complementary to the original databases. For example, the FANTOM web resource aims to present data on the dynamic behavior of transcription and its regulation in the expanding fields of the transcriptome, epigenome and transcriptional networks (14,15). By contrast, this integrated database attaches greater importance to the standardization of data for better distribution, metadata-level integration and cross-database retrieval.
DATABASES TO BE INTEGRATED
In RIKEN, there are a number of databases related to mammalian research resources. In the primary development of the integrated database, we integrated six database projects: the Functional Annotation of the Mammalian Genome 4 (FANTOM 4: http://fantom.gsc.riken.jp) (14–16), the RIKEN Cerebellar Development Transcriptome Database (CDT-DB: http://www.cdtdb.brain.riken.jp/CDT/Top.jsp) (17,18), the resource database from the RIKEN BioResource Center (BRC) (19–21) including mutant resources produced by the ENU mutagenesis program (22,23) and the Resource of Asian Primary Immunodeficiency Diseases (RAPID) (24), the RIKEN Structural Genomics/Proteomics Initiative (RSGI) and two data repositories for the Reference Database of Immune Cells (RefDIC) (25) and the RIKEN Expression Array Database (READ) (26), all of which are produced from individual research projects in the human and mouse. Each database project has its original data schema to represent a variety of data ranging from research resources, such as biological strains, cell lines and DNA clones, to experimental data, such as gene expression and phenotypic analyses. There are no relationships defined among original data tables, which are described by various data formats such as text, images and movies. However, as is usual for most databases, they are compiled in a main data table to represent the objects of the database and related information (Table 1). In the discussions of InterPhenome and CASIMIR, it was recommended that the equivalences or relationships among records from the MGI database for genes and alleles, the International Mouse Strain Resource (IMSR) for experimental strain (27) and terms of OBO ontologies be specified. To show the association between the institute’s data and the public data broadly used in the research community, we constructed an association between RIKEN’s data and public data (Supplementary Table S1).
Table 1.
Imported databases in RIKEN (as for September 2010)
| Database (URL) | Contents | Project URL in SciNetS |
|---|---|---|
| FANTOM4 (http://fantom.gsc.riken.jp/4/) | Monitoring of the dynamics of transcription start site (TSS) usage during a time course of monocytic differentiation in the acute myeloid leukemia cell line THP-1. | http://scinets.org/item/ria187i/ |
| Bio-resource catalog (http://www.brc.riken.jp/) | The online catalog of bioresources including mammalian laboratory strains (mouse), cells and DNA clones in the RIKEN BioResouce Center (BRC). | http://scinets.org/item/ria256i/ |
| RIKEN ENU Mouse Lines (http://www.brc.riken.jp/lab/gsc/mouse/) | Phenotype information of mutant mouse lines generated from large-scale ENU mutagenesis as a resource of the RIKEN BRC. | http://scinets.org/item/rib190i/ |
| Pheno-Pub (http://www.brc.riken.jp/lab/jmc/mouse_clinic/en/m-strain_en.html) | Phenotype data from the standardized phenotyping platform of the Japan Mouse Clinic (JMC) project in the RIKEN BRC. | http://scinets.org/item/ria110i/ |
| Cerebellar Development Transcriptome Database (CDT-DB: http://www.cdtdb.brain.riken.jp/CDT/Top.jsp) | The spatio-temporal gene expression profile of the postnatal development of the mouse cerebellum, | http://scinets.org/item/cria237u1i/ |
| Resource of Asian Primary Immunodeficiency Diseases (RAPID: http://rapid.rcai.riken.jp/RAPID) | A web-based compendium of molecular alterations in primary immunodeficiency diseases. | http://scinets.org/item/cria271u1i/ |
| Systems and Structural Biology Center (SSBC) database (http://www.rsgi.riken.jp/rsgi_e/index.html) | The crystal structures of proteins and the protein–protein interactions in living cells analyzed with the expansion of the genetic code. | http://scinets.org/item/ria46i/ |
| Reference Database of Immune Cells (RefDIC: http://refdic.rcai.riken.jp/welcome.cgi) | An open-access database of quantitative mRNA and protein profiles specifically for immune cells and tissues. | http://scinets.org/item/crib225s27rib225s7i/ |
| RIKEN Expression Array Database (READ: http://read.gsc.riken.jp/) | An integrated system for microarray data that works like ‘glue’ in post-sequence and post-hybridization analyses. | http://scinets.org/item/crib225s27rib225s8i/ |
THE FUNDAMENTALS OF THE INTEGRATED DATABASE: SEMANTIC WEB-BASED CLOUD SYSTEM ‘SCINETS’
We have implemented the integrated database on the data-hosting system, SciNetS, which is a fully web-based common platform that ensures cloud computing in the scientific community on the basis of semantic web technologies (Figure 1). It has multiple features useful for data integration:
The system is designed to support sharing of academic information with secure, and to handle databases for sharing, collaborating or publication. Database developers can set multiple levels of accessibility within user groups or the public for each ‘project’ (private workspace). The user can also declare the copyright licenses for their digital content with Creative Commons (CC) or GNU to indicate the availability for secondary use.
In the project, database developers can also design the semantics with elements and the equivalent methodology to RDF and OWL-Full (i.e. the definition of the semantic links between class and subclass, class and instance, property and sub-property and so on) with graphic user interfaces (GUI) for ontology editors such as Protégé (28). The system assigns the Uniform Resource Identifier (URI) to each data element.
The structured data and metadata can be placed in the public directories of the SciNetS with various standardized data formats, such as RDF, OWL or tab-delimited text file, for downloading or direct connection from external systems and application software such as Protégé.
The system provides the tracking back function making automatically reverse links for RDF relationships across projects. It ensures automatic integration of distributed effort of annotation and curation.
The system is designed to handle a large number of databases simultaneously and is scalable for increased data with the distributed processing technologies on databases and query functions (29). The high-speed retrieval of semantic content is implemented with the General and Rapid Association Study Engine (GRASE), which enables semantic Boolean-based deduction and statistical evaluation of RDF resources (29).
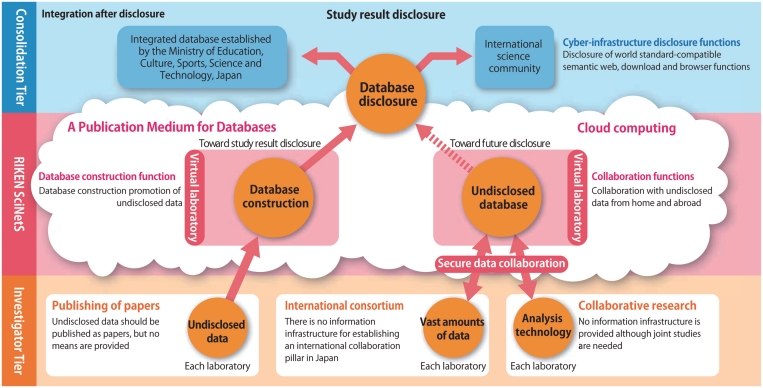
Figure 1.
Schematic diagram showing the concept of SciNetS. SciNetS provides incubation functions from database construction to the integration of databases in computing clouds or a group of large-scale servers, and discloses databases using interfaces compatible with international standards, thus contributing to the establishment of cyber-infrastructure for integrating worldwide databases [reprinted with the courtesy from Tetsuro Toyoda, ‘Synthetic biology—creating biological resources from information resources’ RIKEN RESEARCH 5(10) 13–16, 2010 (http://www.rikenresearch.riken.jp/eng/frontline/6397)].
IMPLEMENTATION OF MAMMALIAN DATA IN SCINETS
The overview of the implementation of this database is presented in Figure 2. The mammalian data and public data shown in Table 1 and Supplementary Table S1, respectively, were imported to SciNetS as individual database projects such that their intact data schema were reflected fully or partially. According to the forms of the original data sources, the databases were imported as three distinct types of projects implemented in SciNetS. First, in the database-type project, a replication of the original database elements, the database table and a data record, is represented with a class and an instance, respectively. Second, the ontology-type project is a replication of the ontology with the OWL methodology. Upon the import of OBO ontologies, ontology files in the OWL format are downloaded from the OBO Foundry website (http://www.obofoundry.org/). Then, the ontology is directly imported into SciNetS. Third, in repository projects, the complete data from a database are stored as single or multiple files. As a result, 27 projects (17 for database, nine for ontology and one for repository) composed of 108 396 classes and 777 319 instances were defined as for September in 2010. These projects are updated monthly in average from constituent databases and ontologies.
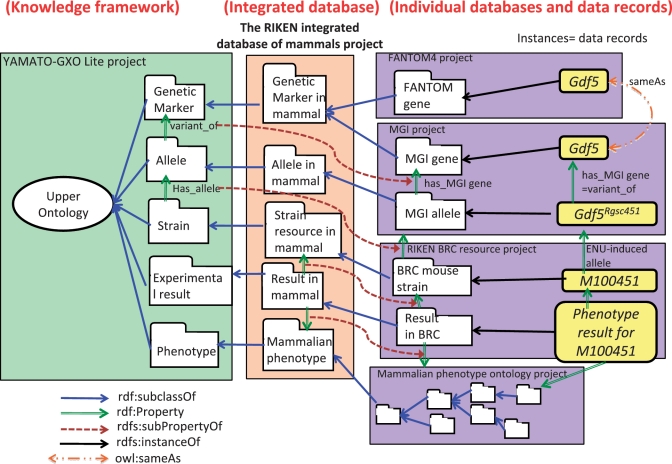
Figure 2.
The implementation of data in the RIKEN integrated database of mammals to ensure direct integration between ontologies and databases based on the semantic web technology. Individual public and RIKEN databases are imported as individual projects, and their main contents are reviewed to classify them into the lower classes to root the 15 classes of the integrated database, such as gene, transcript, experimental data, strain and so on. The classification follows the top-level ontology and is directly linked to the equivalents of rdfs:subclassOf across projects. Property links also organized with rdfs:subPropertyOf to represent the logical definition of upper classes are inherited to lower classes or instances.
Then, we examined the contents and semantics (not the data format or syntax) of 41 classes of imported projects, which play the principal roles in each project. To ensure the consistent classification of the content, we used a top-middle level ontology, YAMATO-GXO Lite (http://scinets.org/item/rib23i/), which is the lightened version of the middle-level ontology, Genetics Ontology (GXO) (30) (http://www.brc.riken.jp/lab/bpmp/ontology/ontology_gxo.html), to bridge between the experimental genetics domain and the latest top-level ontology, Yet Another More Advanced Top-level Ontology (YAMATO) (31) (http://www.ei.sanken.osaka-u.ac.jp/hozo/onto_library/upperOnto.htm). YAMATO-GXO Lite was developed with the ontology editor in SciNetS (paper in preparation). As a result, 41 classes conveying the key information from each project are classified under the fifteen upper classes as follows: ‘Genome segment and gene in mammal’, ‘Allele in mammal’, ‘Transcript in mammal’, ‘Protein in mammal’, ‘Strain resource in mammal’, ‘Cell line resource in mammal’, ‘Disease’, ‘Experimental data with mammalian sample’ and ‘Mammalian Orthologous group’ (Figure 3). The RIKEN Integrated Database of Mammals is implemented as a project to define these classes as a root (http://SciNetS.org/db/mammal). The ontology-based classification of contents was embodied with rdf:subclassOf links, which can be applied across multiple projects in SciNetS. To integrate across species databases, we applied the ‘query-class’, which dynamically refers only to specific instances from another class. For example, the diffraction data class in the SSBC project includes the diffraction data from mammal and non-mammal proteins. To extract only mammal data, we implemented the query-class, which is an expanded use of the owl:oneOf element to define a class by enumerating its elements. With these operations, the project for the integrated database works as the bridge to connect the YAMATO-GXO Lite and imported projects, in which the imported classes are defined as lower concepts of the top-level ontology as shown in Figure 2.
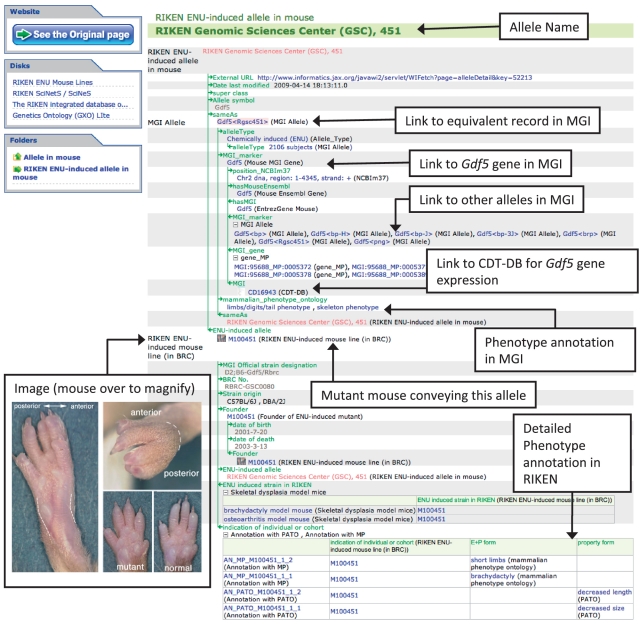
Figure 3.
The instance page to illustrate the graphical representation of detailed semantic links of the RIKEN integrated database of mammals. Explanations are given by open squares with an arrow.
In the next step, to ensure further semantic integration of the imported data, we examined the equivalencies of property links (semantic links) between the upper ontology and lower classes in imported projects. For example, the ‘Allele’ class in YAMATO-GXO Lite has a property named ‘variant_of’ that takes its value from the range of the ‘Genome segment’ class. It is the logical representation of one of the features of an allele that the ‘allele is a variant of a genome segment’. The examination of properties in lower classes reveals that the ‘MGI allele’ class has the ‘MGI gene’ property range of ‘MGI gene’, which is equivalent to ‘variant_of’. Consequently, we defined the ‘MGI gene’ property as a specified type of (rdfs:subPropertyOf) ‘variant_of’ to show that Gdf5Rgsc451, an instance of the MGI allele class, is a variant of Gdf5, an instance of the MGI gene class. With this equivalence mapping of properties between YAMATO-GXO Lite and lower imported database classes, we built the ontology-based information structure so that information defined in the upper classes is instantiated in lower database classes and instances.
In addition, regarding the import of external and internal data records, multiple overlaps of records (instances) were collapsed to represent a single identical entity in the real world (i.e. instances of a gene in the Ensembl, MGI and FANTOM projects). We also examined such equality between instances in lower classes that belong to a single upper class. We related identical data items with a semantic link that is equivalent to owl:sameAs.
USER INTERFACE
At the top page of this integrated database, users can overlook all the classes of integrated databases and those data sizes shown in Supplementary Table S2. The overview of the data structure is presented on the ‘data folder’ page, where users can navigate down the class hierarchy across database or ontology projects by clicking on the folder icons that represent classes. On the page of each project, detailed explanations of the projects and URL links to the original database websites are shown. On the class and instance pages, detailed explanations, a table view of instances, a graphic representation of semantic links and links to original data records are displayed (Figure 3).
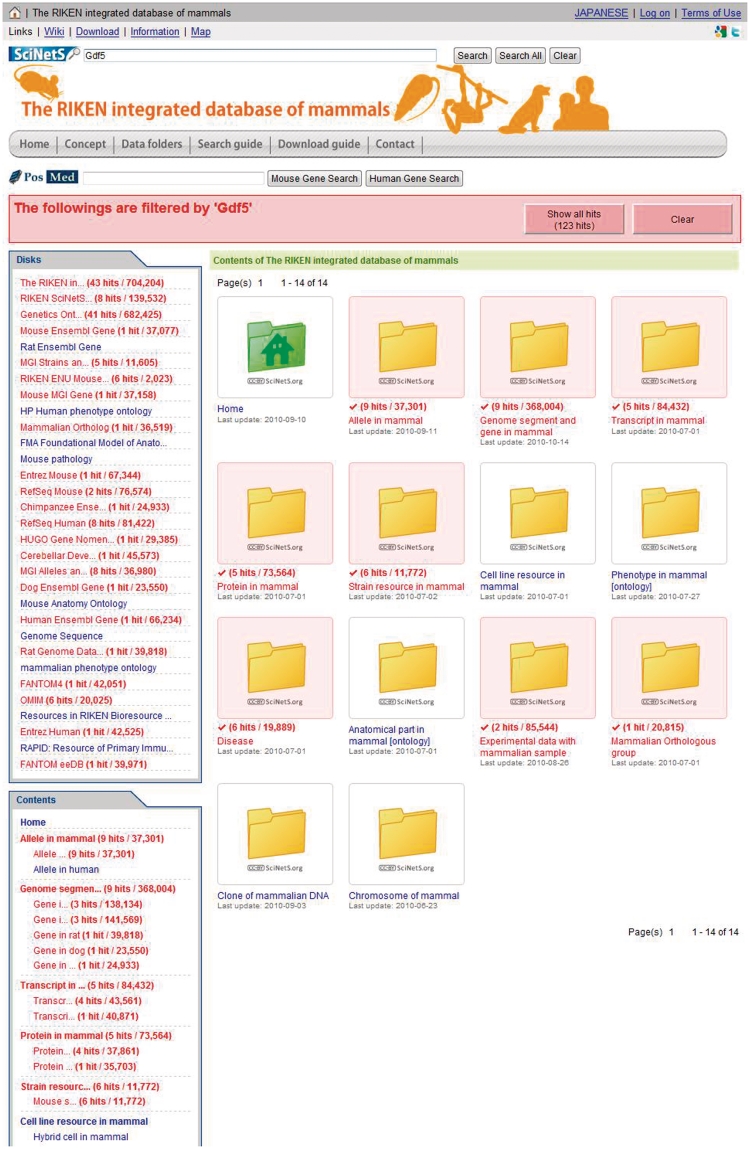
SciNetS implements two kinds of search function, the internal-search and the cross-search. When users search with ‘Search’ button, SciNetS executes internal-search to retrieve queries within the accessing project and related projects and shows the number of query hits on each folder icon of the accessing page (Figure 4). For the cross-search from whole SciNetS data, users can access from ‘Search All’ button. SciNetS replies search results in descending order according to traffic. From this cross-search, users can jump to the Positional Medline (PosMed) search, allows the user to retrieve various information (i.e. gene, phenotypes or diseases) correlated with a genomic position by jumping the PosMed database for a full-document search of various contents: scientific literature, genome annotations, phenome information, protein–protein interactions, co-expression data, orthologous genes, drugs and metabolite information (32,33). These search functions are implemented by our original database search engine GRASE (28). Data in this database are downloadable from the ‘Download’ links of each project with specifications of licenses via CC or GNU. SciNetS provides various several standard formats, such as RDF, OWL or tab-delimited files.
Figure 4.
The representation of filtering (search) result of the RIKEN integrated database of mammals. The number of query hits is represented on each disk or folder icon with red letters.
MERITS OF THE DIRECT INTEGRATION ONTOLOGY WITH DATABASE
The RIKEN integrated database of mammals should be the first practical database to perform the direct integration of the top-level ontology, domain-specific ontologies and the existing databases. Although there is much room for improvement, this database represents a simple and practical methodology to generate a consistent and scalable body of information that is interoperable with the global informational whole based on semantic web technology. In the process of the integration, we have investigated data schema of each database and classified their contents based on the top-level ontology. These operations are comparable to the ‘annotation’ of databases.
Currently, the main knowledge framework is provided by a top-level ontology, YAMATO-GXO lite. During the development of this ontology, it was optimized to allow the integration of multiple biological databases used by the mammalian genetics community. For example, the basic definition of mammalian genes is provided by the Mouse Genomic Nomenclature Committee (MGNC), which is suitable for data management of genome information. It defines gene as ‘a functional unit, usually encoding a protein or RNA, whose inheritance can be followed experimentally’; also, ‘a gene symbol should be unique within the species’. This definition is surely represented in the MGI database because each gene record is stored in the genome segment (phrased as ‘genetic marker’ in MGI) database as a subset (or a subclass) having a biological function and is unique in the mouse genome. An allele is defined as a variant form of a genome segment, which is usually unique for the sequence of itself. Here, we should mention that there are at least two ways to conceptualize genome segments and alleles. One attaches greater importance to the instantiation toward a molecule. Such a classification may be performed in the BioTop top-level ontology (34). Another applies the conceptualization of gene and allele as classes and allows them to have their own instances such as Gdf5 and Gdf5Rgsc451. YAMATO-GXO lite applies latter as useful for integrating databases. A gene is a subclass of the genome segment that has a biological function. An allele is defined as a different class to be unique for conveying information and is equal to the nucleotide sequence.
The consistent knowledge framework contributes to metadata-based and cross-database retrieval for easy and clear specification of the range of the search object. Such retrieval was previously only available for individual databases. For example, to search for ‘the mouse genome segment that has a variant with a point mutation’, a cross-database retrieval is usually performed with the combination of the text, ‘genome segment’ ‘mouse’ and ‘point mutation’. Such a search never indicates the range of the search resource, ‘genome segment of mouse’, which is a subclass of genome segments of mammals. Furthermore, the range must be clearly distinguished from the mouse allele, which is the entity that has the point mutation. In this database, the fifteen upper classes and the lower class-tree are explicitly defined to represent the range of resources and the organization of metadata. Therefore, the knowledge framework enables the retrieval of specific resources, such as ‘genome segment of mouse’, to be related to the text ‘point mutation’ (which may be described in the instance of an allele) using query languages such as SPARQL or GRASQL. On the GUI of this database, the simple GRASQL-based searches are implemented as simple text searches, as described above.
The knowledge framework also contributes to ensuring the cost-effective sustainability and updating of data. In the implementation of SciNetS, the common body for data integration, the continuous maintenance and management of data are essential. These operations are differentiated with respect to not only the formalism of data but also the contents in each database. The consistently integrated data, which represent classification and inheritances between property links, reveal the content-oriented standardization of the formalism of data items. We are now developing content-oriented procedures for data maintenance specified for data contents such as gene, allele and strain. The standardized data formulation provided from top- and middle- level ontologies reduces the labor cost of data management through the reduction of unevenness in the operations of individual databases. Thus, the ‘annotation’ of databases helps to design the contents-oriented common user interfaces or the procedure of data management of imported databases, which had been independently developed in different research projects.
Another advantage of the data integration on SciNetS is that the continuous improvements and enhancements are ensured by the data tracking system to integrate newly added projects. We are planning to incorporate other mammal-related databases into RIKEN to disseminate them to broad communities. Public data are also incorporated to provide higher usability by establishing relationships among data. For example, we still do not ensure fully functional cross-species integration of anatomies and phenotypes, which are provided as species-specific ontologies. To solve this problem, we need equivalence mapping of homologous organs/tissues and phenotypes. Some ontology developers are working on this issue to establish relationships between the Mammalian Phenotype ontology (MP) (35) and Human Phenotype Ontology (HPO) (36–37) mediated by the Phenotypic Quality Ontology (PATO) (38–44). The implementation of such equivalence information in the integrated database will greatly improve the utility of phenotype data to provide cross-mapping information with diseases. Furthermore, we are also integrating the plant omics data using SciNetS with a similar methodology (K. Doi et al. manuscript in preparation). Referring to the same top-level ontology, we are planning to integrate the mammalian database with the plant one. One of the merits of the institute-oriented data integration is the promotion of data integration across phylogenetically distant species because the species- or community-oriented integration of plant and mammal information is often difficult.
FUTURE DIRECTIONS
We will continue the development of this database to enhance the data, retrieval functions and semantics as described above. In addition, we are also planning to incorporate other top-middle level ontologies beyond YAMATO-GXO lite, such as the Basic Formal Ontology (BFO) (45), the Descriptive Ontology for Linguistic, Cognitive Engineering (DOLCE) (46), BioTop and the Ontology of Biomedical Investigation (OBI). In YAMATO, the interoperability among these top-level ontologies represents a general model to explain differentiation and interrelationships among classes (31). With this enhancement, we will cooperate with the global efforts of the OBO Foundry, the initiative activity of the OBO consortium, which has been to coordinate the scientific methods in ontology developments toward forming a consistent, cumulatively expanding and algorithmically tractable whole (7) based on the BFO as the semantic framework.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Maintenance of SciNetS is supported by the Integrated Database Project by Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Drs Kaoru Saijyo, Kazuyuki Mekada and Hatsumi Nakata in RIKEN BRC to help data import from Resource database to SciNetS.
REFERENCES
- 1.Abbott A. Plant genetics database at risk as funds run dry. Nature. 2009;462:258–259. doi: 10.1038/462258b. [DOI] [PubMed] [Google Scholar]
- 2.Maltais LJ, Blake JA, Eppig JT, Davisson MT. Rules and guidelines for mouse gene nomenclature: a condensed version. International Committee on Standardized Genetic Nomenclature for Mice. Genomics. 1997;45:471–476. doi: 10.1006/geno.1997.5010. [DOI] [PubMed] [Google Scholar]
- 3.Wain HM, Lush M, Ducluzeau F, Povey S. Genew: the human gene nomenclature database. Nucleic Acids Res. 2002;30:169–171. doi: 10.1093/nar/30.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. RGD Team. The rat genome database, update 2007–easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouse Phenotype Database Integration Consortium. Hancock JM, Adams NC, Aidinis V, Blake A, Bogue M, Brown SD, Cheslerm EJ, Davidson D, Duran C, et al. Mouse Phenotype Database Integration Consortium: integration of mouse phenome data resources. Mamm. Genome. 2007;18:157–163. doi: 10.1007/s00335-007-9004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandras C, Weaver T, Zouberakis M, Smedley D, Schughart K, Rosenthal N, Hancock JM, Kollias G, Schofield PN, Aidinis V. Models for financial sustainability of biological databases and resources. Database. 2009 doi: 10.1093/database/bap017. doi:10.1093/database/bap017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schofield PN, Bubela T, Weaver T, Portilla L, Brown SD, Hancock JM, Einhorn D, Tocchini-Valentini G, Hrabe de Angelis M, Rosenthal N CASIMIR Rome Meeting participants. Post-publication sharing of data and tools. Nature. 2009;461:171–173. doi: 10.1038/461171a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schatz MC, Langmead B, Salzberg SL. Cloud computing and the DNA data race. Nat. Biotechnol. 2010;28:691–693. doi: 10.1038/nbt0710-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berners-Lee T, Hendler J, Lassila O. Scientific American. 2001. The semantic web. May, pp. 29–37. [Google Scholar]
- 14.FANTOM Consortium. Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaji H, Severin J, Lizio M, Forrest RRA, Nimwegen vE, Rehli M, Shroder K, Irvine K, Susuki H, Carninci P, et al. Update of FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1112. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagami Y, Furuichi T. Investigation of differentially expressed genes during the development of mouse cerebellum. Brain Res. Gene Expr. Patterns. 2001;1:39–59. doi: 10.1016/s1567-133x(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 18.Sato A, Sekine Y, Saruta C, Nishibe H, Morita N, Sato Y, Sadakata T, Shinoda Y, Kojima T, Furuichi T. Cerebellar development transcriptome (CDT-DB): profiling of spatio-temporal gene expression during the postnatal development of mouse cerebellum. Neural Networks. 2008;21:1056–1069. doi: 10.1016/j.neunet.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Yoshiki A, Ike F, Mekada K, Kitaura Y, Nakata H, Hiraiwa N, Mochida K, Ijuin M, Kadotam M, Murakami A, et al. The mouse resources at the RIKEN BioResource center. Exp. Anim. 2009;58:85–96. doi: 10.1538/expanim.58.85. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura Y. Bio-resource of human and animal-derived cell materials. Exp. Anim. 2010;59:1–7. doi: 10.1538/expanim.59.1. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama KK, Murata T, Pan J, Nakade K, Kishikawa S, Ugai H, Kimura M, Kujime Y, Hirose M, Masuzaki S, et al. Genetic materials at the gene engineering division, RIKEN BioResource Center. Exp. Anim. 2010;59:115–124. doi: 10.1538/expanim.59.115. [DOI] [PubMed] [Google Scholar]
- 22.Masuya H, Nakai Y, Motegi H, Niinaya N, Kida Y, Kaneko Y, Aritake H, Suzuki N, Ishii J, Koorikawa K, et al. Development and implementation of a database system to manage a large-scale mouse ENU-mutagenesis program. Mamm. Genome. 2004;15:404–411. doi: 10.1007/s00335-004-2265-8. [DOI] [PubMed] [Google Scholar]
- 23.Masuya H, Yoshikawa S, Heida N, Toyoda T, Wakana S, Shiroishi T. Phenosite: a web database integrating the mouse phenotyping platform and the experimental procedures in mice. J. Bioinform. Comput. Biol. 2007;5:1173–1191. doi: 10.1142/s0219720007003168. [DOI] [PubMed] [Google Scholar]
- 24.Keerthikumar S, Raju R, Kandasamy K, Hijikata A, Ramabadran S, Balakrishnan L, Ahmed M, Rani S, Selvan LD, Somanathan DS, et al. RAPID: Resource of Asian Primary Immunodeficiency Diseases. Nucleic Acids Res. 2009;37:D863–D867. doi: 10.1093/nar/gkn682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hijikata A, Kitamura H, Kimura Y, Yokoyama R, Aiba Y, Bao Y, Fujita S, Hase K, Hori S, Ishii Y, et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23:2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- 26.Bono H, Kasukawa T, Hayashizaki Y, Okazaki Y. READ: RIKEN Expression Array Database. Nucleic Acids Res. 2002;30:211–213. doi: 10.1093/nar/30.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eppig JT, Strivens M. Finding a mouse: the International Mouse Strain Resource (IMSR) Trends Genet. 1999;15:81–82. doi: 10.1016/s0168-9525(98)01665-5. [DOI] [PubMed] [Google Scholar]
- 28.Rubin DL, Noy NF, Musen MA. Protégé: a tool for managing and using terminology in radiology applications. J. Digit. Imaging. 2007;20:34–46. doi: 10.1007/s10278-007-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi N, Toyoda T. Statistical search on the Semantic Web. Bioinformatics. 2008;24:1002–1010. doi: 10.1093/bioinformatics/btn054. [DOI] [PubMed] [Google Scholar]
- 30.Masuya H, Mizoguchi R. Proceedings of the Second Interdisciplinary Ontology Meeting. 2009. Toward fully integration of mouse phenotype information. Keio University Press, February 28–March 1, 2009, Tokyo, Japan, pp. 35–44. [Google Scholar]
- 31.Mizoguchi R. Proceedings of the Second Interdisciplinary Ontology Meeting. 2009. Yet Another Top-level Ontology: YATO. Keio University Press, February 28 - March 1, 2009, Tokyo, Japan, pp. 91–101. [Google Scholar]
- 32.Yoshida Y, Makita Y, Heida N, Asano S, Matsushima A, Ishii M, Mochizuki Y, Masuya H, Wakana S, Kobayashi N, et al. PosMed (Positional Medline): prioritizing genes with an artificial neural network comprising medical documents to accelerate positional cloning. Nucleic Acids Res. 2009;37:W147–W152. doi: 10.1093/nar/gkp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makita Y, Kobayashi N, Mochizuki Y, Yoshida Y, Asano S, Heida N, Deshpande M, Bhatia R, Matsushima A, Ishii M, et al. PosMed-plus: an intelligent search engine that inferentially integrates cross-species information resources for molecular breeding of plants. Plant Cell Physiol. 2009;50:1249–1259. doi: 10.1093/pcp/pcp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz S, Beisswanger E, van den Hoek L, Bodenreider O, van Mulligen EM. Alignment of the UMLS semantic network with BioTop: methodology and assessment. Bioinformatics. 2009;25:i69–i76. doi: 10.1093/bioinformatics/btp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CL, Goldsmith CA, Eppig JT. The mammalian phenotype ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson PN, Mundlos S. The human phenotype ontology Clin. Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 37.Köhler S, Schulz MH, Krawitz P, Bauer S, Dölken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gkoutos GV, Green EC, Mallon AM, Blake A, Greenaway S, Hancock JM, Davidson D. Ontologies for the description of mouse phenotypes. Comp. Funct. Genomics. 2004;5:545–551. doi: 10.1002/cfg.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beck T, Morgan H, Blake A, Wells S, Hancock JM, Mallon AM. Practical application of ontologies to annotate and analyse large scale raw mouse phenotype data. BMC Bioinformatics. 2009;6(Suppl. 5):S2. doi: 10.1186/1471-2105-10-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mungall CJ, Gkoutos GV, Smith CL, Haendel MA, Lewis SE, Ashburner M. Integrating phenotype ontologies across multiple species. Genome Biol. 2010;11:R2. doi: 10.1186/gb-2010-11-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schofield PN, Gkoutos GV, Gruenberger M, Sundberg JP, Hancock JM. Phenotype ontologies for mouse and man: bridging the semantic gap. Dis. Model Mech. 2010;3:281–289. doi: 10.1242/dmm.002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.HancockJ M, Mallon AM, Beck T, Gkoutos GV, Mungall C, Schofield PN. Mouse, man, and meaning: bridging the semantics of mouse phenotype and human disease. Mamm. Genome. 2010;20:457–461. doi: 10.1007/s00335-009-9208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grenon P, Smith B. SNAP and SPAN: towards dynamic spatial ontology. Spat. Cogn. Comput. 2004;4:69–103. [Google Scholar]
- 46.Gangemi A, Guarino N, Masolo C, Oltramari A, Schneider L. Lecture Notes In Computer Science Vol. 2473, 13th International Conference on Knowledge Engineering and Knowledge Management. Ontologies and the Semantic Web. London, UK: Springer; 2002. Sweetening ontologies with DOLCE, knowledge engineering and knowledge management; pp. 166–181. [Google Scholar]