Abstract
This study presents the Yeast Promoter Atlas (YPA, http://ypa.ee.ncku.edu.tw/ or http://ypa.csbb.ntu.edu.tw/) database, which aims to collect comprehensive promoter features in Saccharomyces cerevisiae. YPA integrates nine kinds of promoter features including promoter sequences, genes’ transcription boundaries—transcription start sites (TSSs), five prime untranslated regions (5′-UTRs) and three prime untranslated regions (3′UTRs), TATA boxes, transcription factor binding sites (TFBSs), nucleosome occupancy, DNA bendability, transcription factor (TF) binding, TF knockout expression and TF–TF physical interaction. YPA is designed to present data in a unified manner as many important observations are revealed only when these promoter features are considered altogether. For example, DNA rigidity can prevent nucleosome packaging, thereby making TFBSs in the rigid DNA regions more accessible to TFs. Integrating nucleosome occupancy, DNA bendability, TF binding, TF knockout expression and TFBS data helps to identify which TFBS is actually functional. In YPA, various promoter features can be accessed in a centralized and organized platform. Researchers can easily view if the TFBSs in an interested promoter are occupied by nucleosomes or located in a rigid DNA segment and know if the expression of the downstream gene responds to the knockout of the corresponding TFs. Compared to other established yeast promoter databases, YPA collects not only TFBSs but also many other promoter features to help biologists study transcriptional regulation.
INTRODUCTION
Transcriptional regulation enables a cell to precisely control the temporal and spatial patterns of gene expression in response to physiological or environmental stimuli. The transcription starts from the transcription start site (TSS) and produces a transcript consisting of the 5′-untranslated region (5′-UTR), the coding sequence and the 3′-untranslated region (3′-UTR). In eukaryotes, transcriptional regulation is accomplished by multiple regulatory mechanisms. First, RNA polymerase II must bind to the promoter with the help of TATA binding proteins, which recognize a class of DNA sequences called TATA box, to initialize the transcription. Second, transcription factors (TFs) can modulate the transcription rate. Through binding to specific DNA sequence motifs called transcription factor binding sites (TFBSs) in the promoter, TFs can stabilize or block the binding of RNA polymerase II to the promoter to increase or decrease the transcription rate, respectively. Third, the accessibility of TFBSs to TFs is influenced by the nucleosome positioning. TFBSs occupied by nucleosomes are usually non-functional, presumably because they are not accessible to TFs.
The characteristics of a promoter related to these regulatory mechanisms, such as the presence/absence of TATA box, are named ‘promoter features’ in this study. Most promoter features have been shown in yeast to be correlated with each other. For example, TFBSs are found to be enriched in the region of 100–200 bp upstream from the TSS (1). In TATA box-less promoters, such TFBS-enriched region is shown to favor nucleosome-depleted and rigid DNA regions in the promoter (2–4). Tirosh and Barkai (3) also observed that the presence/absence of nucleosomes in the proximity of TSS influences the transcriptional plasticity—the capacity to modulate gene expression upon changing conditions.
The complicated connections among different promoter features emerge an immediate need to construct a promoter database that integrates comprehensive promoter features to help biologists study transcriptional regulation. Many promoter features in Saccharomyces cerevisiae have been extensively studied. However, these valuable resources were scattered over literature, supplementary data and even authors’ personal homepages. Most existing yeast promoter databases—e.g. TRANSFAC (5), SCPD (6), JASPAR (7), YEASTRACT (8), SwissRegulon (9), MYBS (10), TransfactomeDB (11)—were designed to deposit experimentally verified or computationally predicted TFBS data. In this article, we present the Yeast Promoter Atlas (YPA) database, which aims at collecting many promoter features related to the transcriptional regulation and providing an interface to query and browse these data simultaneously. The promoter features collected in YPA includes promoter sequences, genomic locations of TSSs, 5′-UTRs, 3′-UTRs, TATA boxes and TFBSs, TF binding, TF knockout expression and TF–TF physical interactions. The nucleosome occupancy and bendability at every base pair in the yeast genome are also provided. By integrating TFBSs with many other promoter features, YPA helps biologists to pick up functional TFBSs with multiple lines of evidence. In YPA, the collected promoter features are represented in three forms. The first form is a tabulated view providing the most comprehensive information; the second form is an interactive genome browser; the third view is a plain text for downloading and further manipulation. Moreover, YPA is designed to present data in a unified manner as many important observations are revealed only when these promoter features are considered altogether.
DATA COLLECTION
Our database collects nine kinds of promoter features from six databases and five articles. First, the DNA sequences of the yeast genome were downloaded from the SGD database (12). Second, the genomic locations of the start and stop codons of 6603 genes and the TSSs, 5′-UTRs and 3′-UTRs of 4560 genes were retrieved from Nagalakshmi et al.’s work (13). Third, the genomic locations of 2983 TATA boxes in the promoters of 2115 genes were retrieved from Basehoar et al.’s work (4). The fourth promoter feature is the genomic locations of TFBSs of 164 TFs collected from five sources. The first source is the article of MacIsaac et al. (14), which used PhyloCon and Converge algorithms to predict TFBSs of 112 TFs. The second source is the MYBS database (10), which collected TFBSs of 72 TFs from 11 TFBS databases. The third source is the SwissRegulon database (9), which used Phylogibbs algorithm to predict TFBSs of 79 TFs. The fourth and fifth sources are the YEASTRACT (8) and SCPD databases (6), which collected experimentally verified TFBSs of 102 and 27 TFs, respectively. These TFBSs were obtained from published footprinting or ChIP experiments (6,8).
Fifth, the nucleosome occupancy at every base pair in the yeast genome was retrieved from Kaplan et al.’s work (15). The nucleosome occupancy at every base pair is calculated as the log-ratio between the number of reads that cover that base pair and the average number of reads per base pair. Sixth, the bending propensity of each tri-nucleotide was retrieved from Brukner et al.’s work (16). They used the DNase I digestion to estimate the bending propensity. Seventh, the binding evidences of 25 180 TF–promoter pairs based on band-shift, footprinting or ChIP assays in literature were retrieved from the YEASTRACT database (8). It can tell us whether a specific TF can bind to a target promoter. Eighth, the regulation evidences of 19 090 TF–gene pairs based on TF knockout assays in literature were retrieved from the YEASTRACT database (8). It can tell us whether the expression of a target gene would change significantly in response to a specific TF knockout. Finally, 409 TF–TF physical interactions with experimental evidence were retrieved from the BioGRID database (17).
DATA PROCESSING
In this database, the promoter region of a gene is defined as the intergenic region from the start codon of this gene to the coding region boundary of its nearest non-overlapped upstream gene. For all regulatory elements (TFBSs and TATA boxes) located in the promoter region of a gene, we calculated their relative distances from the TSS (or the start codon if the TSS is not available). Besides, the TFBS data collected from the five sources were refined to 19 TFBS datasets under different parameter settings. In MacIsaac et al.’s article (14), two motif discovery algorithms were used to identify the TFBSs present in the promoter regions of genes bound by the same TF (determined by the P-value of the ChIP-chip data) with a phylogenetic conservation constraint (requiring the same binding sites present in the orthologous promoter regions of phylogenetically related yeast species). Different ChIP-chip P-value thresholds and phylogenetic conservation constraints result in different TFBS datasets. In MYBS database (10), ChIP-chip P-value and phylogenetic conservation were also applied as two filters to select TFBSs. Therefore, by using two ChIP-chip P-values (0.001 and 0.005) and three phylogenetic conservation constraints (conserved in one, two and three yeast species), YPA generates 12 different TFBS datasets (six from MacIsaac et al. and six from MYBS). In SwissRegulon database (9), the putative genome-wide locations of 79 TFBSs were reported with a posterior probability. Over 85 000 locations are recorded, of which approximately 57 000 have a posterior probability >0.1 and approximately 17 000 have a posterior probability >0.5. In YPA, we provide five cutoffs (0.1, 0.2, 0.3, 0.4 and 0.5) for the posterior probability, thus generating five different TFBS datasets. The last two TFBS data sets are those TFBSs located in the promoter regions (defined by YPA) retrieved from the YEASTRACT (8) and SCPD (6) databases, respectively.
The original data of nucleosome occupancy of each base pair is a real number (15). YPA sets a threshold of this nucleosome occupancy to define the nucleosome-occupied region. A base pair is said to be in a nucleosome-occupied region if its nucleosome occupancy is within the highest K% of those in the yeast genome, where K = 10–90 is chosen by users. The bending propensity reported in Brukner et al’s article (16) is a table of 64 values associating with 64 tri-nucleotide tuples. We first assigned the i-th base pair a raw bendability with the propensity corresponding to the tuple of (i–1, i, i+1), the i-th base pair and its two flanking ones. This raw bendability was then smoothed with a moving average of 31 base pairs. After obtaining a real number representing the bendability at each base pair, we adopted a criterion to define the rigid DNA region similar to the definition of nucleosome-occupied region. A base pair is said to be in a rigid DNA region if its DNA bendability is within the lowest K% among of those in the yeast genome, where K = 10–90 is chosen by users.
DATABASE INTERFACE
The YPA database consists of three major views: (i) a comprehensive page displaying all information collected/processed by YPA for a specific promoter region, (ii) an interactive genome browser providing the neighboring information of a genomic coordinate and (iii) a plain-text representation containing the corresponding information of (i) and (ii) for downloading and further manipulation. The homepage of YPA provides entry points to the three major views and to a configuration page. The users of YPA can configure all the parameters described in the ‘DATA PROCESSING’ section in the configuration page. Following are the descriptions of the three views.
Promoter page
Users can specify a precise gene/ORF name, e.g. RPL23A or YBL087C, or a keyword including wild card characters, e.g. RPL*, to query the database. YPA also allows users to (i) search for the genes whose promoters contain the user-specified TFBSs and (ii) search for the TFBSs that are in the promoters of the user-specified genes.
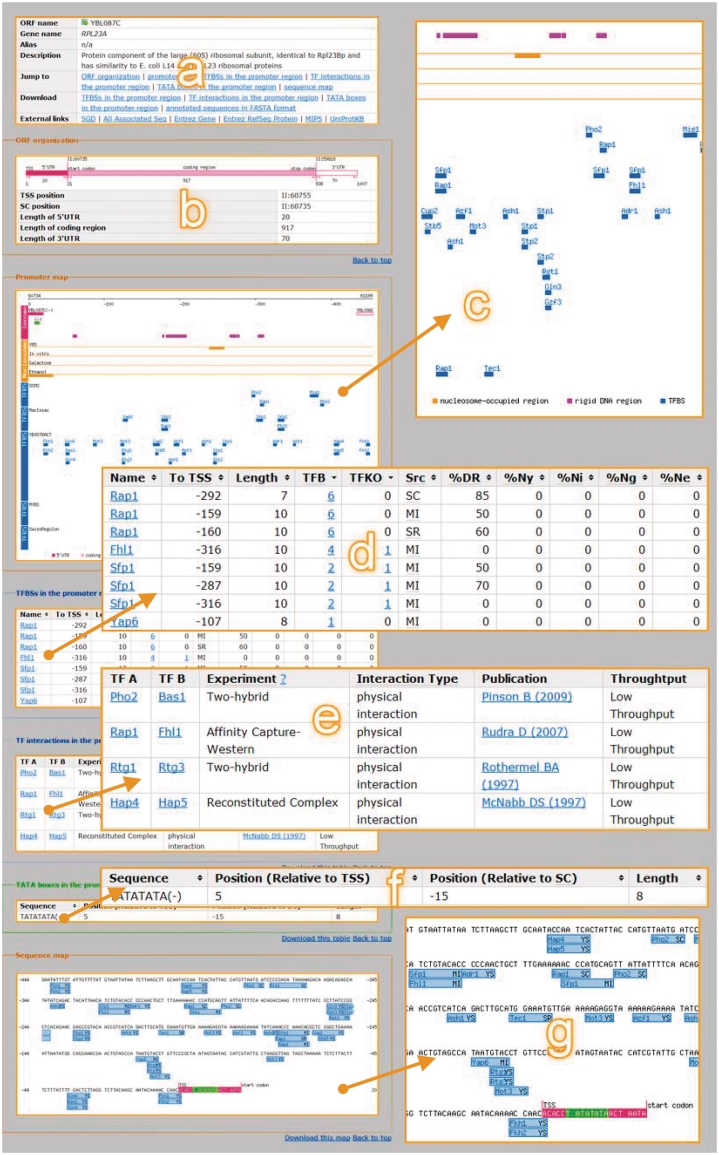
Here we use RPL23A as an example (Figure 1) to demonstrate the usage of the promoter page. The promoter page consists of seven areas. The first area (Figure 1a) provides the basic information, including the name, descriptions and related external links of the gene/ORF. The second area (Figure 1b) displays the ORF organization, including the genomic locations of the coding region and, if available, of the 5′-UTR and 3′-UTR. The third area (Figure 1c) is a map plotting the regulatory elements in the promoter region of the queried gene. The fourth area is a table (Figure 1d) listing all the TFBSs in the promoter region of the queried gene. The information of each TFBS includes the TF name, the relative distances from the TSS or the start codon, the length of the TFBS, the number of references providing the TF binding and TF regulation evidences and the proportions of the nucleotides of the TFBS located in the rigid and nucleosome-occupied DNA regions. The fifth area (Figure 1e) shows the physical interactions between two TFs that have TFBSs in this promoter region. The sixth area (Figure 1f) lists the TATA boxes in this promoter region. The seventh area (Figure 1g) is a nucleotide-level map where users can read the sequence.
Figure 1.
The promoter page of RPL23A in YPA.
Genome browser
The second major view of YPA is an interactive genome browser. Users can specify a genomic coordinate, e.g. II:60k, to invoke the genome browser. Users can zoom in and out and scroll through the genome in the browser. After reaching a satisfactory view, a text file containing all the information of the browsed region is downloadable. This facility allows users to retrieve data containing multiple promoter regions.
Plain-text representation
YPA provides a text representation of a specific genomic range in FASTA format allowing users to download for further manipulation. The first sequence in our downloadable FASTA file is the whole sequence of the retrieved range while the remaining sequences are ORFs, TFBSs and TATA boxes within the range.
CASE STUDY
We use RPL23A (Figure 1) as an example to demonstrate the advantage of displaying various promoter features simultaneously. Figure 1c shows 44 TFBSs of 26 TFs in the promoter region of RPL23A collected in YPA. As most of these TFBSs were identified in silico, biologists have to resort to other evidence to figure out the most confidential ones to perform further analyses. Among these predicted TFBSs, the binding sites of Rap1, Fhl1 and Sfp1 are most likely to be functional. Several lines of evidence support this assertion (Table 1). First, ChIP-chip experiments show that Rap1, Fhl1 and Sfp1 can bind to the promoter of RPL23A (Figure 1d). Second, TF knockout microarrays show that Fhl1 and Sfp1 can regulate the expression of RPL23A (Figure 1d). Third, most binding sites of Rap1 and Sfp1 are located in the rigid DNA region and nucleosome-free region, making these TFBSs more accessible to the TFs (Figure 1d). Finally, affinity capture-Western experiments show that Rap1 and Fhl1 have physical interaction (Figure 1e). Our speculation is supported by the literature. The protein product of RPL23A is a ribosomal protein and Rap1, Fhl1 and Sfp1 are three major TFs known to regulate genes that encode ribosomal proteins (12,18–20).
Table 1.
Promoter features that strengthen the confidence of the three computationally predicted TFBSs in the promoter region of RPL23A
| Evidence from promoter feature | Rap1 | Fhl1 | Sfp1 |
|---|---|---|---|
| Evidence of the TF binding to this promoter | v | v | v |
| Evidence of the significant expression change of the TF knockout | v | v | |
| TF–TF physical interaction | v | v | |
| Located in a nucleosome-free region | v | v | v |
| Located in a rigid DNA region | v | v |
COMPARISON WITH EXISTING PRMOTER DATABASES
There have been a number of databases providing the information related to the yeast promoters. Most of them aim at collecting TFBS data. SCPD focuses on experimentally determined TFBSs (6). MYBS collects computationally and experimentally determined TFBSs for several yeast species (10). TRANSFAC is a commercial database providing high-quality TFBSs of many organisms (5). SwissRegulon deposits TFBSs of 17 prokaryotes and three eukaroytes (9). JASPAR database contains binding profiles derived from published collections of experimentally defined TFBSs (7). Some databases provide auxiliary data in addition to the TFBS data. YEASTRACT collects TF–gene regulatory relations from literatures (8). TransfactomeDB provides condition-specific regulatory information for TFBSs (11). SGD provides the richest yeast information (12). However, a large part of SGD information is less related to the promoter features, such as protein sequences, molecular functions, subcellular localizations and literature information.
Compared to the databases described above, the TF binding and TF knockout expression data are only available in YEASTACT and YPA; the TF–TF interaction data are only available in SGD and YPA; while the genes’ transcription boundaries and DNA bendability are only available in YPA. Furthermore, YPA’s data are highly integrated. For example, when users want to know whether two TFs having TFBSs in the same promoter region have physical interaction, they only need to query YPA once, but have to query SGD several times for their genomic locations and interaction data individually. Table 2 summarizes the promoter features collected in current yeast promoter databases in the public domain. Note that this table focuses on YPA’s features. It is used to demonstrate the uniqueness of YPA but not to prove that YPA is superior over other databases. By collecting the richest promoter information and providing a highly integrated platform, YPA aims to make these regulatory annotations more informative than just available. In addition, we provide a text representation that is easy for parsing and analyzing computationally. Through the friendly interface, the YPA resource will be useful for people studying transcriptional regulation both experimentally and computationally.
Table 2.
Promoter features collected in YPA and seven yeast promoter databases in the public domain
| Promoter feature | YPA | SGD | SCPD | SwissRegulon | MYBS | YEASTRACT | TransfactomeDB | JASPAR |
|---|---|---|---|---|---|---|---|---|
| TFBSa | v | v | v | v | v | v | v | v |
| TFBS locationb | v | v | v | v | v | v | ||
| TATA boxc | v | v | v | v | v | |||
| TATA box locationd | v | v | v | |||||
| TSS location | v | v | ||||||
| 5′- and 3′-UTR | v | |||||||
| TF binding | v | v | ||||||
| TF knockout expression | v | v | ||||||
| TF–TF physical interaction | v | v | ||||||
| Nucleosome occupancy | v | v | ||||||
| DNA bendability | v |
aThe database provides TFBS consensus, weight matrix or sequence logo.
bThe database provides the genomic location of TFBSs.
cThe database provides the sequence of TATA boxes.
dThe database provides the genomic location of TATA boxes.
However, YPA has two limitations. First, YPA collects TFBSs of only 164 yeast TFs. Although this collection is more comprehensive than other existing TFBS databases, YPA does not cover all yeast TFs. The reason is that high-confidence TFBSs for the remaining TFs are still lacking. Second, all of YPA’s data were collected from published resources rather than newly generated or predicted by YPA. Although the data integration in YPA may provide some knowledge, literature support or further experiments are still needed.
CONCLUSION
In this article, we present the YPA database that focuses on yeast promoters. YPA provides an easy-to-use interface for browsing the yeast genome and viewing multiple promoter features simultaneously. Researchers can retrieve the data collected from various sources with a few operations. YPA will be regularly updated based on the newest release of the related databases and from newly published literature. Furthermore, to exploit novel connections among available promoter features deserves further studies in the future.
FUNDING
Funding for open access charge: National Science Council Taiwan (NSC 99-2628-B-006-015-MY3 and NSC 99-2628-E-006-017).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The TF binding and TF regulation evidences collected from the YEASTRACT database provide convincing clues for users to assess whether a given TFBS is likely to be functional. We greatly appreciate the effort of the YEASTRACT team in collecting such valuable data. We also want to thank Dr. Huang-Mo Sung for the helpful comments.
REFERENCES
- 1.Lin Z, Wu WS, Liang H, Woo Y, Li WH. The spatial distribution of cis regulatory elements in yeast promoters and its implications for transcriptional regulation. BMC Genomics. 2010;11:581. doi: 10.1186/1471-2164-11-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tirosh I, Berman J, Barkai N. The pattern and evolution of yeast promoter bendability. Trends Genet. 2007;23:318–321. doi: 10.1016/j.tig.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 5.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
- 7.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteiro PT, Mendes ND, Teixeira MC, d'Orey S, Tenreiro S, Mira NP, Pais H, Francisco AP, Carvalho AM, Lourenço AB, et al. YEASTRACT-DISCOVERER: new tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:D132–D136. doi: 10.1093/nar/gkm976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai HK, Chou MY, Shih CH, Huang GT, Chang TH, Li WH. MYBS: a comprehensive web server for mining transcription factor binding sites in yeast. Nucleic Acids Res. 2007;35:W221–W226. doi: 10.1093/nar/gkm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foat BC, Tepper RG, Bussemaker HJ. TransfactomeDB: a resource for exploring the nucleotide sequence specificity and condition-specific regulatory activity of trans-acting factors. Nucleic Acids Res. 2008;36:D125–D131. doi: 10.1093/nar/gkm828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macisaac KD, Wang T, Gordon BD, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brukner I, Sánchez R, Suck D, Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995;14:1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, Warner JR. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell Biol. 2006;26:4853–4862. doi: 10.1128/MCB.02367-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 20.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl Acad. Sci. USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]