Abstract
ODB (Operon DataBase) aims to collect data of all known and conserved operons in completely sequenced genomes. Three newly updated features of this database have been added as follows: (i) Data from included operons were updated. The genome-wide analysis of transcription and transcriptional units has become popular recently and ODB successfully integrates these high-throughput operon data, including genome-wide transcriptional units of five prokaryotes and two eukaryotes. The current version of our database contains information from about 10 000 known operons in more than 50 genomes, and more than 400 000 conserved operons obtained from more than 1000 bacterial genomes. (ii) ODB proposes the idea of reference operons as a new operon prediction tool. A reference operon, a set of possible orthologous genes that organize operons, is defined by clustering all known operons. A large number of known operons, including the recently added genome-wide analysis of operons, allowed us to define more reliable reference operons. (iii) ODB also provides new graphical interfaces. One is for comparative analyses of operon structures in multiple genomes. The other is for visualization of possible operons in multiple genomes obtained from the reference operons. The 2011 updated version of ODB is now available at http://operondb.jp/.
INTRODUCTION
In the field of functional genomics, operon information is often used to observe associations between genomes and biological functions. Genes in an operon are often functionally related hence they appear in the same biological pathway and work together as one system. Today, the genomic data of more than 1000 species are available and the next-generation sequencing techniques should provide an unprecedented amount of genomes. In order to extract biological knowledge from a large amount of genomic and metagenomic data, the standard and most helpful technique is to utilize the genomic contexts such as operons and transcriptional units.
We have constructed ODB, the Operon DataBase, which is a collection of operons and transcriptional units documented in the literature and putative operons that are conserved in terms of the known operons (1). Today, a number of databases have been developed to address operons (1–6); however, ODB is the first database that integrates known operons comprehensively in multiple genomes described in the literature and newly identified operons detected by high-throughput transcriptomic analyses have been added in this update. Recently, global transcription analysis techniques such as RNA-seq and tiling arrays have been developed and details of transcriptional maps in some genomes have been revealed (7–14). The genome-wide transcriptional map allows us to know the global organization of transcriptional units. The current version of ODB includes transcriptional unit data derived from seven genome-wide transcriptional maps, and the number of operons and transcriptional units stored in ODB is now about 10 000. As far as we know, ODB is the most abundant source of operons and transcriptional units, which have all been examined in the literature and/or wet-experiments.
In addition, a large number of known operon data enabled us to extract putative operons conserved in multiple genomes. Some of these putative operons are often functionally or evolutionally associated. Regarding them as a group of operons, ODB provides a newly developed tool to predict operons from the orthologous genes of the group. We have recently improved functionality and visualization interfaces for the comparison of operon organizations. The recent update of ODB is described in the following section.
OPERON DATA SOURCES
In the 2011 update, ODB contains about 10 000 known operons and transcriptional units from more than 50 organisms (Table 1). These known operons increase the database to about five times more than the previous version of ODB. We collected information about these known operons and transcriptional units from studies in the literature, which are based on several wet-experiments ranging from direct methods such as primer extensions and northern blots to less direct methods such as gene knock-out experiments. The analyses of genome-wide transcription and transcriptional units in some genomes have been recently performed. In these studies, the transcription was investigated throughout a whole genome by tiling array techniques, and the regions of possible transcriptional units were defined. We also added these genome-wide transcriptional units from Escherichia coli (8), Helicobacter pylori (10), Listeria monocytogenes (12), Mycoplasma pneumonia (9), Sulfolobus solfataricus (13) in prokaryotes and Caenorhabditis elegans (7) and Ciona intestinalis (14) in eukaryotes. In the previous version of ODB, we included only eukaryotic operon information from C. elegans, and in the current version of ODB we added the large-scale operon information from C. intestinalis and one operon from Drosophila melanogaster. It is still unclear why some eukaryotic genomes organize operons, however, from the viewpoint of the comparison of transcriptional organizations between prokaryotic and eukaryotic genomes, we have added the information of eukaryotic operons.
Table 1.
Statistics of known and predicted operons and transcriptional units stored in ODB
| Number of operons | Number of genes | Number of genomes | |
|---|---|---|---|
| Known operon | 9890 | 23 415 | 56 |
| Predicted operon | 415 193 | 649 314 | 1136 |
An operon found in an organism is often conserved in other organisms. However, the gene order in an operon is often shuffled and collapsed in its evolutionary history (15), hence the gene order of the conserved operon is not always same as the original order. These conserved operons are expected to be strongly functionally related. Thus, we searched operons conserved in multiple genomes using the known operon as a query. The data of orthologous genes were obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG) Ortholog Cluter (OC) (ftp://ftp.genome.jp/pub/kegg/genes/oc/oc.gz) (16), which is a set of orthologous genes clustered computationally. If genes in a known operon have orthologous genes in other genome and these orthologous genes are consecutively located on the same strand of the genome, we regarded them as a new conserved operon. Therefore, if their consecutiveness is interrupted by other genes, they were not regarded as new conserved operons. As the result of this search, we found about 400 000 conserved operons in more than 1000 genomes (Table 1). Both the numbers of genomes and known operons were increased by about five times more than the numbers in the previous version, hence the conserved operons increased by 30 times.
NEW OPERON PREDICTION
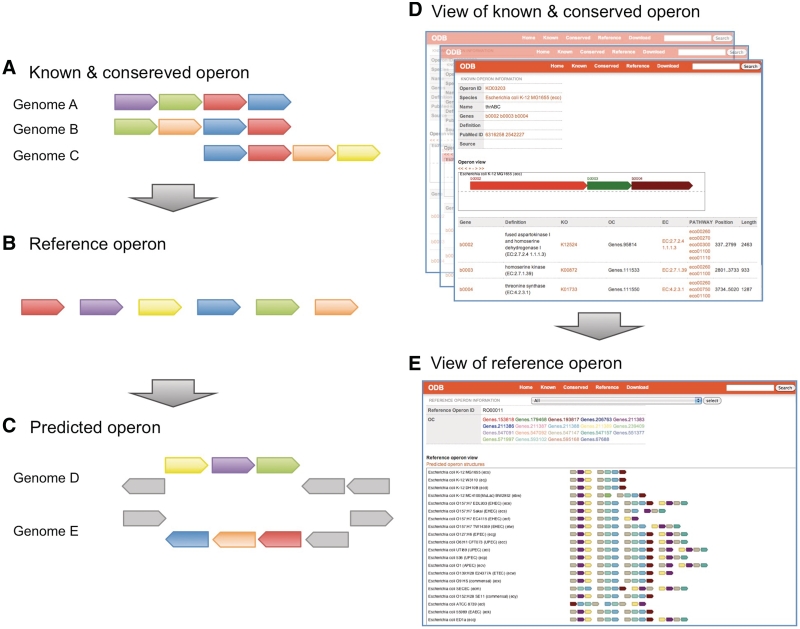
ODB provides a new tool to predict operons based on the idea of reference operons. A reference operon is a set of possible orthologous genes that organize operons. Some operons are often conserved across genomes, but the contents of the genes are not always the same. In this case, common genes in multiple operons are well conserved and are thought to be strong functionally related. On the other hand, the other genes seem to be less functionally related compared to the common genes, but they are important for organizing an operon. The idea of a reference operon is to integrate such operons so that a part of them overlaps (Figure 1). Although this idea is similar to ‘uber-operons,’ (17,18) we call them ‘reference operons,’ because we can predict possible operons in a genome by mapping orthologous genes on a reference operon, in the same way as orthologous genes are mapped on the ‘reference pathway’ maps in the KEGG PATHWAY (16). A large number of known operons, including recently updated operons identified by transcriptomic analyses, allowed us to define more reliable reference operons. The reference operons are defined by clustering all known operons we have collected. We used the Markov Clustering algorithm (MCL) (19) with inflation parameter 6 for constructing reference operons. As a result, we were able to successfully define 4812 reference operons. Thus, a reference operon is a set of all orthologous gene identifiers from known operons and transcriptional units that overlap. The current version of ODB includes more than 1000 genomes, so we can easily predict reliable operon structures in these genomes. On the other hand, this method is not used for the genome-wide operon prediction such as the genome context method or machine learning method. Because this method is based on known operon structures and their conservation, the coverage of predicted operon structures in a genome by this method is limited. However, we think that this issue will be resolved in the near future by collecting more known operon information from the literature and recent global transcription analyses such as the genome-wide tiling array and RNA-seq analysis can accelerate the defining of our reference operons.
Figure 1.
Contents of the ODB database. (A) Known and conserved operon structures. (B) An orthologous gene set of a refrence operon obtained from (A). (C) Putative operon structures predicted by mapping (B) to genomes. (D and E) Screenshots of web interfaces of known and conserved operons and reference operons.
COMPARATIVE GENOMIC VIEWER
ODB provides a web interface to view operon information and the organizations in genomes. When users search a keyword of interest for genes or operons from the search box at the top right corner of the page, the summary list of the retrieved information related to the operon is shown. The detailed information page is linked from each identifier of known/conserved/reference operons on the retrieved summary list as matched to the query keyword. In known and conserved operons, the genomic viewer allows observation of the operon structure and the peripheral genes, scrolling and zooming into the region of interest on the genome. The user is also provided with a graphical interface to compare the conserved operon structures across multiple genomes by selecting a taxonomic level. All information including graphic symbols is linked to the KEGG database or other suitable database if available.
By accessing the web page for a reference operon, users can retrieve the list of orthologous genes included in the reference operon. By selecting a taxonomic level, users can be presented with a view that compares the predicted operon structures across multiple genomes within the selected taxonomic level (Figure 1). In addition, users can retrieve the orthologous table that indicates the presence or absence of the orthologous genes in each genome.
DATA DOWNLOAD AND ACCESSIBILITY
ODB also provides a data downloading system. Users can download all information of known and conserved operons that we have collected and the reference operons. Users can access ODB at the URL (http://www.genome.sk.ritsumei.ac.jp/odb2/), and also at the URL (http://operondb.jp/) as a simple URL for entry into this database. The previous version of ODB is also provided at the URL (http://www.genome.sk.ritsumei.ac.jp/odb/).
FUTURE PERSPECTIVES
The relationship between consecutive gene transcription via operon and transcriptional units, and discrete gene transcription via a transcriptional factor, remains unclear. To understand the differences in the transcriptional system, including operons and regulons, we need more accurate and genome-wide operon maps. In the near future, more global transcription analysis should drastically enrich the information of operons and transcriptional units. This expanded knowledge of operons should allow us to improve our understanding of transcriptional systems.
FUNDING
Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). Funding for open access charge: grant-in-aid for Young Scientists (B).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Yuki Moriya and Susumu Goto for the technical support of ODB.
REFERENCES
- 1.Okuda S, Katayama T, Kawashima S, Goto S, Kanehisa M. ODB: a database of operons accumulating known operons across multiple genomes. Nucleic Acids Res. 2006;34:D358–D362. doi: 10.1093/nar/gkj037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010;38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao FL, Dam P, Chou J, Olman V, Xu Y. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 2009;37:D459–D463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009;37:D479–D482. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- 8.Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 10.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 11.Sorek R, Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 12.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 13.Wurtzel O, Sapra R, Chen F, Zhu YW, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, et al. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol. 2008;9:R152. doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Che DS, Li GJ, Mao FL, Wu HW, Xu Y. Detecting uber-operons in prokaryotic genomes. Nucleic Acids Res. 2006;34:2418–2427. doi: 10.1093/nar/gkl294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lathe WC, Snel B, Bork P. Gene context conservation of a higher order than operons. Trends Biochem. Sci. 2000;25:474–479. doi: 10.1016/s0968-0004(00)01663-7. [DOI] [PubMed] [Google Scholar]
- 19.van Dongen S. 2000. Graph clustering by flow simulation. Ph.D. Thesis, University of Utrecht. [Google Scholar]