Abstract
ModBase (http://salilab.org/modbase) is a database of annotated comparative protein structure models. The models are calculated by ModPipe, an automated modeling pipeline that relies primarily on Modeller for fold assignment, sequence–structure alignment, model building and model assessment (http://salilab.org/modeller/). ModBase currently contains 10 355 444 reliable models for domains in 2 421 920 unique protein sequences. ModBase allows users to update comparative models on demand, and request modeling of additional sequences through an interface to the ModWeb modeling server (http://salilab.org/modweb). ModBase models are available through the ModBase interface as well as the Protein Model Portal (http://www.proteinmodelportal.org/). Recently developed associated resources include the SALIGN server for multiple sequence and structure alignment (http://salilab.org/salign), the ModEval server for predicting the accuracy of protein structure models (http://salilab.org/modeval), the PCSS server for predicting which peptides bind to a given protein (http://salilab.org/pcss) and the FoXS server for calculating and fitting Small Angle X-ray Scattering profiles (http://salilab.org/foxs).
INTRODUCTION
Genome sequencing efforts are providing us with complete genetic blueprints for hundreds of organisms. We are faced with assigning and understanding the functions of proteins encoded by these genomes. This task is generally facilitated by knowing the proteins’ 3D structures, which are best determined by experimental methods such as X-ray crystallography and NMR spectroscopy. In the last two years, the number of experimentally determined protein structures in the Protein Data Bank (PDB) has increased by 30% to 67 794 (September 2010) (1). However, in the same timeframe, the number of protein sequences in the comprehensive public sequence databases such as GenBank (2) and UniProtKB (3) has grown even more rapidly; for example, the number of sequences in UniProtKB has nearly doubled to >12 million. Protein structure prediction methods are attempting to bridge this gap. The need for accurate models can sometimes be met by homology or comparative modeling (4–8). Comparative modeling is carried out in four sequential steps: identifying known structures (templates) related to the sequence to be modeled (target), aligning the target sequence with the templates, building models and assessing the models. For this reason, comparative modeling is only applicable when the target sequence is detectably related to a known protein structure.
As more experimental structures become available, and more reliable models become accessible to the biologists, web-accessible resources that assist in analyzing protein structures and structural models and evaluating their reliability become of increasing importance.
Here, we describe the current state of the ModBase database of comparative protein structure models, the ModWeb comparative modeling web-server and several new associated resources: the SALIGN server for multiple sequence and structure alignment (http://salilab.org/salign) (9), the ModEval server for predicting the accuracy of protein structure models (http://salilab.org/modeval), the PCSS server for predicting which peptides bind to a given protein (http://salilab.org/pcss) (10) and the FoXS server for calculating and fitting Small Angle X-ray Scattering profiles (http://salilab.org/foxs) (11). We also present new modules of the UCSF Chimera molecular graphics package that retrieve models from ModBase and act as a graphical interface to Modeller. Finally, we illustrate the use of comparative models by calculating modeling leverage for structural genomics, superfamily member identification and functional annotation, prediction of protein–protein interactions and genome-wide functional annotation.
CONTENTS
Model generation by comparative modeling (Modeller and ModPipe)
Models in ModBase are calculated using our automated software pipeline for comparative protein structure modeling, ModPipe (12). The software relies mostly on modules of Modeller (13), and is designed to process data sets of protein sequences on a Linux cluster.
ModPipe uses sequence–sequence (14), sequence–profile (7,15) and profile–profile (7,16) methods for fold assignment and target–template alignment, using a promiscuous E-value threshold of 1.0 to increase the likelihood of identifying the best available template structure. These alignments can cover only a segment or the whole target sequence. By default, for each target–template alignment, 10 models are calculated (13) and the model with the best value of the Discrete Optimized Protein Energy (DOPE) statistical potential (17) is selected and then evaluated by several additional quality criteria: (i) target–template sequence identity, (ii) GA341 score (18), (iii) Z-DOPE score (17), (iv) ModPipe Quality Score (MPQS) and (v) TSVMod score (19). The MPQS score is a composite model quality criterion that includes the coverage of the modeled sequence, sequence identity, the fraction of gaps in the alignment, the compactness of the model and various statistical potential Z-scores. A short description of the other scores can be found below in the section ‘ModEval: server for predicting errors in structural models’. The models that score best with at least one of these quality criteria are selected for further filtering. If more than 30 residues of a target sequence are not covered by a selected model, additional models are selected even if they don’t score best with at least one of the quality criteria. Finally, only the models with quality criteria values above specified thresholds or with an E-value <10−4 are included in the final model set.
A key feature of the pipeline is not prejudging the validity of sequence–structure relationships at the fold-assignment stage; instead, sequence–structure matches are assessed after the construction of the models and their evaluation. This approach enables a thorough exploration of fold assignments, sequence–structure alignments and conformations, with the aim of finding the model with the best evaluation score, at the expense of increasing the computational time significantly, since for some sequences, a few thousand models can be calculated.
The source code for ModPipe is freely accessible under the GPL terms (http://salilab.org/modpipe). The binary code for Modeller is also available freely to academics for a number of different machine types (http://salilab.org/modeller).
ModBase model sets
Models in ModBase are organized in data sets. Because of the rapid growth of the public sequence databases, we concentrate our efforts on adding data sets that are useful for specific projects, rather than attempt to model all known protein sequences with detectable template structures. Currently, ModBase includes a model data set for each of 43 complete genomes, as well as a data set for the complete SwissProt/TrEMBL database (2005) (http://salilab.org/modbase/statistics). We identified the genomes with the highest access statistics (Homo sapiens, Saccharomyces cerevisiae, Escherichia coli, Mycobacterium tuberculosis, Mus musculus, Arabidopsis thaliana, Drosophila melanogaster, Rattus norvegicus and Caenorhabditis elegans), and are updating the corresponding models more frequently (approximately once a year). Together with other project-oriented data sets, ModBase currently contains 10 355 444 reliable models for domains in 2 421 920 unique sequences.
ModWeb: comparative modeling web-server
The ModWeb comparative modeling web-server is an integral module of ModBase (http://salilab.org/modweb) (12). In the default mode, ModWeb accepts one or more sequences in the FASTA format, followed by calculating and evaluating their models using ModPipe based on the best available templates from the PDB. Alternatively, ModWeb also accepts a protein structure as input, calculates a multiple sequence profile and identifies all homologous sequences in the UniProtKB database, followed by modeling these homologs based on the user-provided structure. This alternative protocol is a useful tool for measuring the impact of new structures, such as those generated by structural genomics efforts (20). Additionally, new members of sequence superfamilies with at least one known structure can be identified (21).
In addition to the existing anonymous access, we recently added a user registration option. Registered users get unified access to all their ModWeb data sets and can submit template-based calculations.
ASSOCIATED RESOURCES
A number of web-services are associated with ModBase. Some of these are tightly integrated with ModBase, while others contain data that are derived through ModBase—e.g. single nucleotide polymorphism (SNP) annotations created by LS-SNP (22). We have already described the interactions of ModBase with the ModLoop server for loop modeling in protein structures (http://salilab.org/modloop) (23), the PIBASE database of protein–protein interaction (http://salilab.org/pibase) (24), the DBAli database of structural alignments (http://salilab.org/dbali) (25,26) and the LS-SNP database of structural annotations of human non-synonymous single-nucleotide polymorphisms (http://salilab.org/LS-SNP) elsewhere (22,27,28). Here, we describe several additional servers that are now interacting with ModBase.
SALIGN: server for multiple sequence and structure alignment
Accurate alignment of protein sequences and structures is crucial for comparative modeling; for example, sequence–structure alignment is needed for template identification (16) and target–template alignment (29); structure–structure alignments are useful for comparing multiple templates with each other (9), in preparation for comparative modeling based on multiple template structures (13). The SALIGN web-server (http://salilab.org/salign) performs sequence–sequence, sequence–structure and structure–structure alignments of two or more proteins (H. Braberg et al., manuscript in preparation). Depending on the provided input and desired output, a number of different algorithms and options implemented in Modeller can be applied, including global and local dynamic programming; linear and non-linear gap penalty functions; sequence- and structure-based similarity matrices and progressive/tree-based multiple alignments (9,16,29,30).
Given an input of sequences and/or structures, the server proposes the optimal alignment protocol. For instance, given more than two input structures and sequences, the structures and sequences are separately aligned to each other. The two multiple alignments are then aligned with one another, making use of the variable gap penalty function (29). Two sets of multiple sequence alignments can also be aligned using a profile–profile method (16). The user can override the default choice of algorithms and parameters. We have previously demonstrated the effectiveness of the algorithms used in the server in the context of comparative modeling (28,31) and identification of interacting protein partners (32).
ModEval: server for predicting errors in structural models
Model evaluation is an essential step in protein structure modeling, as its results allow the user to judge the level of accuracy of the model and whether or not a model is suitable for the intended application. Two model evaluation methods are available within Modeller. First, GA341 (18) is a statistical potential-based score, which discriminates between models of correct and incorrect fold. It is derived from a nonlinear combination (evolved by a genetic algorithm) of three model features (33): model length, ZPAIR (a distance statistical potential Z-score) and ZSURF (a surface-accessibility statistical potential Z-score). The two Z-scores are combined in the ZCOMB score. Second, the DOPE score is an atomic-distance-dependent statistical potential derived from known protein structures (17). To facilitate comparison between models of different sequences, a normalized DOPE score (Z-DOPE) for the whole model is also reported, as is a profile of the residue Z-DOPE scores that allows identification of problematic regions of a model.
Recently, we developed TSVMod (19,34), a method to estimate the Cα RMSD error and the native overlap (the fraction of Cα atoms within 3.5 Å of their native positions) of a model. The error prediction relies on a model-specific scoring function constructed by a support vector machine that optimizes the weights of up to nine features, including various sequence similarity measures and statistical potentials, extracted from a tailored training set of models unique to the model being assessed. If possible, the training relies on similarly sized models with the same fold; otherwise, similarly sized models with the same secondary structure composition are used.
The ModEval server (http://salilab.org/modeval) accepts a protein structure, an alignment in the PIR format (optional) and the sequence–template sequence identity (optional). It then computes the TSVMod scores, the Z-DOPE score and profile and all components of the GA341 score. Upon completion of the job, the user receives an email notification.
PCSS: server for predicting which peptides bind to a given protein
Protein–protein recognition is frequently mediated by small peptide regions of one protein binding to a pocket or groove of another protein. Examples include scaffolding domains such as PDZ and SH3 (35), which recognize peptides 6–10 residues in length; and protease–substrate specificity, in which the substrate peptide associates with the protease active site cleft before catalysis (36). This recognition is mediated by the sequence of the peptide and its structural environment in the binding protein. It is often helpful to be able to identify these peptides; for example, detecting a peptide that is cleaved by a protease can lead to hypotheses of the effect of this cleavage on protein substrate function. To aid in this prediction effort, the PCSS web-server (http://salilab.org/pcss) has been created that allows the user to provide positive and negative examples of peptide binding to a given protein. From these training data, a statistical model is generated that can then be used by the server to search for similar peptides in other protein sequences.
The PCSS web-server has two modes, ‘Training’ and ‘Application’. In the training mode, the user uploads a set of proteins containing the peptides of interest, specified by their UniProtKB accession numbers. The user indicates for each peptide whether it is a positive or negative example of the peptide motif. The server then validates the input and uses the sequence and structure features of the peptides to create a support vector machine model. The structure features of the peptides are derived from experimental structures or high-quality comparative models in ModBase, when available. In the application mode, the user provides a set of target proteins and uses the model created in the training mode to search for further examples of positive peptides. While training support vector machines generally requires expert knowledge, the PCSS server automates the process of feature selection and encoding, parameter sampling and benchmarking, thereby increasing the efficiency of its construction.
The algorithm implemented in PCSS was recently used to predict two substrates of the pro-apoptotic serine protease Granzyme B (GrB) (10): apoptosis-inducing factor 1 and survival motor neuron protein 1. Both were experimentally validated as being a GrB substrate in vitro, and are implicated in apoptosis. Their cleavage potentially represents a mechanism that natural killer cells and cytotoxic lymphocytes use to induce programmed cell death in virally-infected and neoplastic cells.
FoXS: server for calculating and fitting Small Angle X-ray Scattering profiles
Small Angle X-ray Scattering (SAXS) is a common technique for low-resolution structural characterization of molecules in solution (37–39). SAXS experiments determine the scattering intensity of a molecule as a function of spatial frequency, resulting in a SAXS profile that can be easily converted into the approximate distribution of atomic distances in the measured system. SAXS experiments can be performed with the protein sample in solution, and usually take only a few minutes on a well-equipped synchrotron beamline (39).
FoXS (http://salilab.org/foxs) is a rapid and accurate method for calculating a SAXS profile of a given molecular structure based on the Debye formula (11). The method explicitly computes all inter-atomic distances, and models the first solvation layer based on solvent accessibility. FoXS was tested with all eight structures in the PDB that have an experimental SAXS profile in the open access SAXS database (http://bioisis.net/) as well as 16 additional structures with SAXS profiles from our collaborations. The FoXS resource can contribute to many applications, such as comparing a conformation in solution with the corresponding X-ray structure, modeling a flexible or multi-modular protein and assembling a macromolecular complex from its subunits.
VISUALIZATION AND ANALYSIS OF ALIGNMENTS AND MODELS WITH CHIMERA
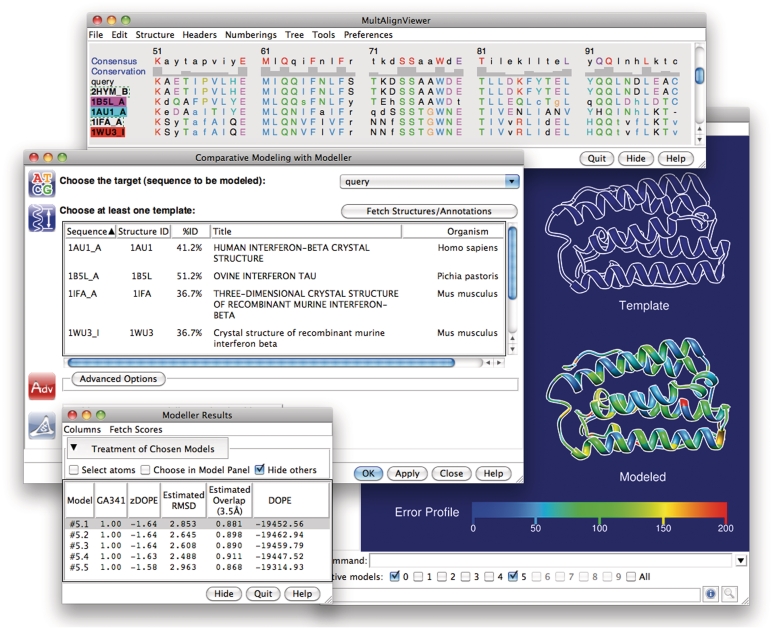
UCSF Chimera is a graphics program for analysis and interactive visualization of molecular structures and related data (40). New modules have been added to Chimera for interaction with ModBase and Modeller. From within Chimera, all models for a given sequence in ModBase can be retrieved over the web by entering a sequence identifier (such as the UniProtKB accession number) into the Chimera ‘Fetch by ID’ dialog or command line. The fetched models are displayed in the main Chimera window, and their scores, residue range, template identifier and other information are listed in a table (similar to Figure 1, bottom left). Any of the general analysis features in Chimera can be applied to the models, such as calculation of hydrogen bonds, steric clashes and structure superpositions. The PDB files returned by ModBase contain content to allow for coloring the model by the degree to which the restraints have been satisfied, which can be used to predict model errors (Figure 1, right).
Figure 1.
The Chimera–Modeller interface. The sequence alignment is displayed in Chimera's Multalign Viewer tool (top). In the dialog for running Modeller (middle left), one of the sequences in the alignment is designated as the target, and at least one structure (associated with another sequence in the alignment) is designated as the template. Structure information is shown to help guide the choice of template. After the run, the resulting models are listed along with various model scores from Modeller in a table (bottom left) and their structures are loaded into Chimera. In this example, the main Chimera window (right) shows the template as an outline and one of the model structures as a ribbon colored by error profile.
Additional new functionality in UCSF Chimera includes a graphical interface to build a model from scratch using Modeller, using as input only the amino acid sequence of the target protein. Chimera uses BLAST to search the PDB for potential templates, which are displayed in the Multalign Viewer tool (Figure 1, top) (41). The Viewer allows for alignment editing, for example, to remove gaps that fall within an element of regular secondary structure in the template, which frequently contribute to model error. Additional sequences can be added to the alignment, either as text or from other structures in Chimera. When the alignment is satisfactory, the user builds models using Modeller within Chimera. This process is run in the background and can be monitored via Chimera’s task manager. When the results become available, the models are displayed in Chimera and their scores shown in a table (Figure 1, bottom left). This functionality is also available for models already stored in ModBase, to allow for refinement of those models through editing the alignment and incorporating additional templates. Chimera can run a locally installed copy of Modeller or use a Modeller web service provided by the UCSF Resource for Biocomputing, Visualization, and Informatics (http://www.rbvi.ucsf.edu).
Model assessment by interactive visualization of structures and template–target sequence alignments is an important complement to the statistical scores available in ModBase. While model evaluation scores allow efficient filtering of the models most likely to be correct (17,19), interactive visualization may better reveal specific problematic regions, and more importantly, may allow for adjusting such regions in an iterative alignment/modeling process.
APPLICATION EXAMPLES
Modeling leverage for structural genomics: a BenF-like porin from Pseudomonas fluorescens
One of the metrics guiding target selection in structural genomics is modeling leverage. Modeling leverage of a structure is defined as the number of proteins sequences that can be modeled based on the structure at >30% sequence identity. The New York Structural GenomiX Research Center (NYSGXRC) recently determined the structure of a putative BenF-like porin from P. fluorescens (PflBenF), which has the same fold as structurally defined members of the OprD superfamily (20). Members of this superfamily are thought to mediate transport of most small molecules across the cell membrane in Pseudomonads (42). To determine the modeling leverage of PflBenF, template-based modeling as implemented in ModWeb was performed, using the sequences and structures of PflBenF as well as two previously determined similar structures, OpdK (43) and OprD (44), both from P. aeruginosa. A total of 221 unique protein sequences were identified in the UniProtKB database, with sequence identities >30% to at least one of these three protein structures. The first structure of a member of this fold family, PaOprD, enabled modeling of 165 related proteins. Subsequent determination of the structure of PaOpdK resulted in models for an additional three protein sequences. In contrast, determination of the PflBenF structure enabled homology modeling of 53 additional protein sequences. Thus, the structure of PflBenF expands significantly the number of useful homology models of the porins in the OprD and OpdK families. Experimental structures of additional OprD/OpdK subfamily members should provide useful guides for planning experiments aimed at defining the mechanisms governing pore selectivity. The modeling leverage statistics for this project can be accessed at http:///modbase.compbio.ucsf.edu/modbase-cgi/model_leverage.cgi?type=master_partha.
Superfamily member identification and functional annotation: Solute Carrier Transporters
Solute carriers are a group of approximately 400 biomedically important membrane proteins that control the uptake and efflux of solutes, including essential cellular compounds and therapeutic drugs (45). Numerous variants that are important for clinical drug response have been identified in solute carriers by the Pharmacogenomics of Membrane Transporters project (PMT) at UCSF (46). Solute carriers can share similar structural features despite weak sequence similarities.
We defined solute carrier families by comparing their sequences using structure and profile–profile alignments as well as similarity networks. The families were analyzed in the context of substrate type, transport mode, organism conservation and tissue specificity (47). The classification is useful for inferring similarities and differences in various structural and functional features such as fold, ligand-binding site and molecular mechanism of uncharacterized solute carriers based on their characterized aligned homologs. We used these family definitions to show which solute carriers have known structures or have good quality comparative models—i.e. models based on >30% sequence identity to a known template structure over at least 70% of their sequences, or are assessed to have the correct fold by various scores (47). In addition to ModBase and the Protein Model Portal (48), the solute carrier alignments and models are freely accessible via PharmGKB (49). A phylogenetic tree for each modeled solute carrier is also provided through a link from the ModBase model pages (http://salilab.org/modbase/search?dataset=slc).
Prediction of protein–protein interactions: Schistosoma mansoni pathogenesis
S. mansoni is a parasitic flatworm and the major causative agent of schistosomiasis, a disease affecting >200 million people in developing countries. The pathogen employs many strategies to infect the human host and evade the immune response through different life-cycle stages (50). To understand these mechanisms of pathogenesis, we applied a host–pathogen protein–protein interaction prediction pipeline to the human and S. mansoni proteomes. This pipeline, previously applied on 10 pathogens (51), relies on comparative modeling of human and pathogen proteins based on template domain–domain interactions and subsequent evaluation of the complex model interface using the MODTIE statistical potential (32). Application of the pipeline resulted in over 500 predicted complexes involving both human and S. mansoni proteins. Some of these predictions include parasite proteins expressed in the invasive cercarial life-cycle as well as human proteins known to play a role in immunomodulatory processes. Several of these predictions are currently being tested by experiment.
Genome-wide functional annotation: the Helicobacter pylori proteome
The Gram-negative bacterium H. pylori inhabits the human stomach. The presence of pathogenic strains has been shown to lead to gastric ulcers, gastritis and gastric cancer (52). As part of our effort to provide functional annotations for genes in the H. pylori genome (http://phylogenomics.berkeley.edu/phylofacts/), we created a ModBase data set of models for all sequences in the proteome of the H. pylori strain 26695 that are detectably related to an experimental structure. For 61 of the 1575 proteins in this strain, crystal structures of domains or whole proteins already exist. For 1467 of the remaining 1514 proteins in this strain, at least one reliable model was built. The number of proteins with models based on 0–20, 20–30, 30–40, 40–50, 50–60 and 60–100% sequence identity is 40, 368, 603, 275, 96 and 85, respectively. Of these, 584 had at least one model for which TSVMod (19) predicted a Cα RMSD ≤ 3.5 Å. The available templates lie at varying evolutionary distances from the target proteins, and different regions of a single target protein may be homologous to different templates.
We illustrate the use of these models with the enzyme biotin carboxylase (locus HP_0370, UniProt accession O25134, gi 2313468). Biotin carboxylase catalyzes an early step in fatty acid biosynthesis. Thus, bacterial biotin carboxylases are investigated as potential drug targets using virtual screening (53). Because these enzymes occur across the Tree of Life (including human), detailed knowledge of the catalytic site geometry may help in designing drugs that are specific to the pathogen and don’t bind to the host proteins. Prediction of functional sites by similarity to experimentally characterized functional sites is facilitated by the use of comparative models to visualize and probe protein function (25,54,55).
The ModPipe pipeline produced several models for this protein based on templates at different evolutionary distances. Analysis of the H. pylori biotin carboxylase with the Berkeley PHOG algorithm (56), a phylogenomic method of orthology prediction, supports the annotation of this protein as a biotin carboxylase based on super-orthology—the most stringent definition of orthology (57)—with two experimentally characterized proteins in the BRENDA database (58): Q54755 (Synechococcus elongatus strain PCC 7942) and Q10YA8 (Trichodesmium erythraeum strain IMS101). A human mitochondrial ortholog, Q96RQ3 (PDB ID 2ejm), includes annotation of site-specific features from SwissProt (59).
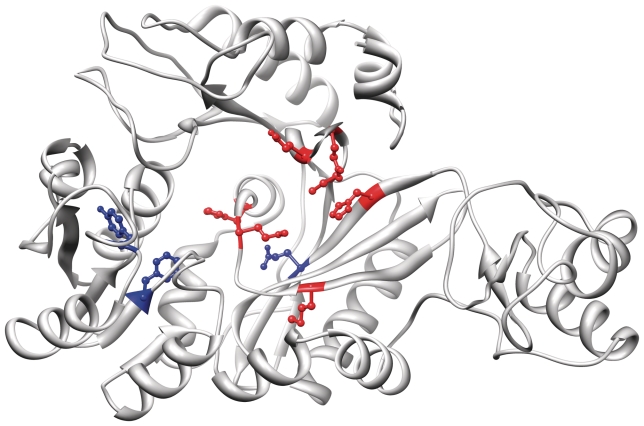
To predict functional residues using the ModBase models for this enzyme, we submitted the H. pylori biotin carboxylase to the INTREPID webserver (60) that uses a phylogenomic algorithm to predict evolutionarily conserved sites (61). Of the top 10 residues predicted by INTREPID, 7 are supported by experimental studies based on homology to the biotin carboxylase subunit of Acetyl-Coa Carboxylase (PDB ID 1bnc): C243 [equivalent to C230 in 1bnc, whose catalytic function is supported (62)], H222 [H209 in 1bnc (63)], H312 [H297 in 1bnc, adjacent to active site (63)], M304 [M289 in 1bnc (63)], Q246 [Q233 in 1bnc (64)], Q250 [Q237 in 1bnc (63)] and Q309 [Q294 in 1bnc (63)]. Three residues (F93, Y74 and Q226) may represent novel predictions of functional sites. INTREPID predictions and known active site residues are displayed in Figure 2, illustrating the use of comparative models to predict functional sites. The complete genome modeling data set for H. pylori can be downloaded from ftp://salilab.org/databases/modbase/projects/genomes/.
Figure 2.
Model of Helicobacter pylori biotin carboxylase based on template 1dv1. TSVMod predicts a Cα RMSD of 3.5 Å. The top ten functional residues predicted by INTREPID are highlighted: seven that are also known from the literature to be involved in catalytic function are colored red, and three representing potential novel predictions are colored blue. These 10 residues are, in descending order of INTREPID importance score: C243 (red), H222 (red), H312 (red), F93 (blue), M304 (red), Y74 (blue), Q226 (blue), Q246 (red), Q250 (red) and Q309 (red). UCSF Chimera was used to load the model from ModBase and produce this figure.
ACCESS AND INTERFACE
Direct access
The main access to ModBase is through its web interface at http://salilab.org/modbase, by querying with UniprotKB (3) and GI (2) identifiers, gene names, annotation keywords, PDB (65) codes, data set names, organism names, sequence similarity to the modeled sequences (BLAST (15)) and model-specific criteria such as model reliability, model size and target–template sequence identity. Additionally, it is possible to retrieve coordinate files and alignment files as text files. Select genome data sets are also available from our ftp server (ftp://salilab.org/databases/modbase/projects).
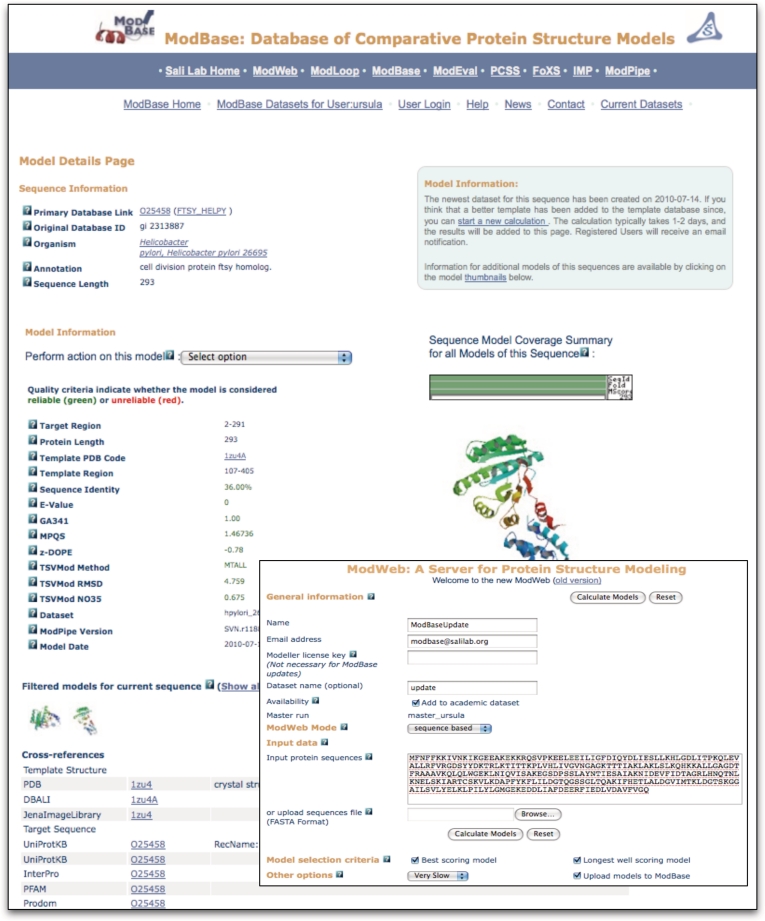
The output of a search is displayed on pages with varying amounts of information about the modeled sequences, template structures, alignments and functional annotations. An example of the output from a search resulting in one model is shown in Figure 3. A ribbon diagram of the model with the highest target–template sequence identity is displayed by default, together with some details of the modeling calculation. Ribbon thumbprints of additional models for this sequence link to corresponding pages with more information. Ribbon diagrams are generated on the fly using Molscript (66) and Raster3D (67). A pull-down menu provides links to additional functionality: the SNP module, retrieval of coordinate and alignment files as well as molecular visualization by Chimera that allows the user to display template and model coordinates together with their alignment. If mutation information is available for a protein sequence, links to the details are provided in the cross-references section. Additionally, cross-references to various other databases, including PDB (65), UniProtKB (68), the UCSC Genome Browser (69), EBI’s InterPro (70), PharmGKB (71) and SFLD (72) are given. Other ModBase pages provide overviews of more than one sequence or structure. All ModBase pages are interconnected to facilitate easy navigation between different views.
Figure 3.
ModBase Model Details page (e.g. O25458 from the Helicobacter pylori genome data set): Prominently displayed is the model with the highest sequence identity/model length combination. The thumbprints represent all models from the most recent modeling calculation. Models from earlier calculations are also available. A ribbon diagram of the primary model, database annotations, and modeling details are displayed. The pull-down menu provides access to alternative ModBase views and other types of information (if available), such as data about SNPs. The cross-references section contains links to relevant internal and external databases. Through a link to ModWeb (displayed in the inner box), a user can update the model.
Access through external databases
The Protein Model Portal (PMP) has become a valuable option for accessing ModBase models (http://proteinmodelportal.org) (49,73). The PMP is a single point of entry for accessing protein structure models from a number of different databases, by querying all participating source model databases, and serving the model coordinates, alignments and quality criteria from a central location.
ModBase models in academic and public data sets are also directly accessible from several other databases, including UniProtKB (3), PIR’s iProClass (68), EBI’s InterPro (70), the UCSC Genome Browser (69), PubMed (LinkOut) (74), PharmGKB (71) and SFLD (72).
FUTURE DIRECTIONS
ModBase will grow by adding models calculated on demand by external users (using ModWeb) as well as our own calculations of model data sets that are needed for our research projects (using ModPipe, ModWeb or Modeller). These updates will reflect improvements in the methods and software used for calculating the models as well as new template structures in the PDB and new sequences in UniProtKB. In the future, we expect that most of the users will access ModBase models through the PMP.
CITATION
Users of ModBase are requested to cite this article in their publications.
FUNDING
National Institutes of Health (R01 GM54762, U54 GM074945, U54 GM074929, U01 GM61390, P01 GM71790 to A.S., F32 GM088991 to A.Sch., P41 RR001081 to T.E.F.); the National Science Foundation (0732065 to A.S. and K.S.); the Department of Energy (DE-SC0004916 to K.S.); Sandler Family Supporting Foundation (to A.S). Funding for open access charge: NIH (U54 GM074945).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
For linking to ModBase from their databases, the authors thank Torsten Schwede (PMP), David Haussler and Jim Kent (UCSC Genome Browser), Amos Bairoch (SwissProt/TrEMBL), Rolf Apweiler (InterPro), Patsy Babbitt (SFLD), Russ Altman (PharmGKB) and Kathy Wu (PIR/iProClass). The authors are also grateful for computing hardware gifts from Mike Homer, Ron Conway, NetApp, IBM, Hewlett Packard and Intel.
REFERENCES
- 1.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 5.Wallner B, Elofsson A. All are not equal: a benchmark of different homology modeling programs. Protein Sci. 2005;14:1315–1327. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillisch A, Pineda LF, Hilgenfeld R. Utility of homology models in the drug discovery process. Drug Discov. Today. 2004;9:659–669. doi: 10.1016/S1359-6446(04)03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eswar N, Webb B, Marti-Renom M, Madhusudhan M, Eramian D, Shen M, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Prot. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 9.Madhusudhan MS, Webb BM, Marti-Renom MA, Eswar N, Sali A. Alignment of multiple protein structures based on sequence and structure features. Protein Eng. Des. Sel. 2009;22:569–574. doi: 10.1093/protein/gzp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkan D, Hostetter D, Mahrus S, Pieper U, Wells J, Craik C, Sali A. Prediction of protease substrates using sequence and structure features. Bioinformatics. 2010;26:1714–1722. doi: 10.1093/bioinformatics/btq267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneidman-Duhovny D, Hammel A, Sali A. FoXS: a web server for rapid computation and fitting of saxs profiles. Nucleic Acids Res. 2010;38:541–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eswar N, John B, Mirkovic N, Fiser A, Ilyin VA, Pieper U, Stuart AC, Marti-Renom MA, Madhusudhan MS, Yerkovich B, et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 14.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marti-Renom MA, Madhusudhan MS, Sali A. Alignment of protein sequences by their profiles. Protein Sci. 2004;13:1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melo F, Sali A. Fold assessment for comparative protein structure modeling. Protein Sci. 2007;16:2412–2426. doi: 10.1110/ps.072895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eramian D, Eswar N, Shen M, Sali A. How well can the accuracy of comparative protein structure models be predicted? Protein Sci. 2008;17:1881–1893. doi: 10.1110/ps.036061.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampathkumar P, Lu F, Zhao X, Li Z, Gilmore J, Bain K, Rutter ME, Gheyi T, Schwinn K, Bonanno J, et al. Structure of a putative BenF-like porin from Pseudomanas fluorescens Pf-5 at 2.6Å resolution. Prot. Struct. Func. Bioinform. 2010;78:3056–3062. doi: 10.1002/prot.22829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pieper U, Chiang R, Seffernick J, Brown S, Glasner M, Kelly L, Eswar N, Sauder J, Bonanno J, Swaminathan S, et al. Target selection and annotation for the structural genomics of the amidohydrolase and enolase superfamilies. J. Struct. Funct. Genom. 2009;10:107–125. doi: 10.1007/s10969-008-9056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 23.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 24.Davis F, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 25.Marti-Renom MA, Pieper U, Madhusudhan MS, Rossi A, Eswar N, Davis FP, Al-Shahrour F, Dopazo J, Sali A. DBAli tools: mining the protein structure space. Nucleic Acids Res. 2007;35:W393–W397. doi: 10.1093/nar/gkm236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marti-Renom MA, Ilyin VA, Sali A. DBAli: a database of protein structure alignments. Bioinformatics. 2001;17:746–747. doi: 10.1093/bioinformatics/17.8.746. [DOI] [PubMed] [Google Scholar]
- 27.Pieper U, Eswar N, Webb B, Eramian D, Kelly L, Barkan D, Carter H, Mankoo P, Karchin R, Marti-Renom M, et al. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madhusudhan MS, Marti-Renom MA, Sanchez R, Sali A. Variable gap penalty for protein sequence-structure alignment. Protein Eng. Des. Selec. 2006;19:129–133. doi: 10.1093/protein/gzj005. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Z, Sali A, Blundell T. A variable gap penalty function and feature weights for protein 3-D structure comparisons. Protein Eng. 1992;5:43–51. doi: 10.1093/protein/5.1.43. [DOI] [PubMed] [Google Scholar]
- 31.Pieper U, Eswar N, Braberg H, Madhusudhan MS, Davis FP, Stuart AC, Mirkovic N, Rossi A, Marti-Renom MA, Fiser A, et al. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2004;32:D217–D222. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis FP, Braberg H, Shen MY, Pieper U, Sali A, Madhusudhan MS. Protein complex compositions predicted by structural similarity. Nucleic Acids Res. 2006;34:2943–2952. doi: 10.1093/nar/gkl353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melo F, Sanchez R, Sali A. Statistical potentials for fold assessment. Protein Sci. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eramian D, Shen M, Devos D, Melo F, Sali A, Marti-Renom M. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko T, Li L, Li SS. The SH3 domain—a family of versatile peptide—and protein-recognition module. Front Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 36.Pardo J, Aguilo JI, Anel A, Martin P, Joeckel L, Borner C, Wallich R, Mullbacher A, Froelich CJ, Simon MM. The biology of cytotoxic cell granule exocytosis pathway: granzymes have evolved to induce cell death and inflammation. Microbes Infect. 2009;11:452–459. doi: 10.1016/j.micinf.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 39.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence–structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Hancock RE, Brinkman FS. Function of pseudomonas porins in uptake and efflux. Ann. Rev. Microbiol. 2002;56:17–38. doi: 10.1146/annurev.micro.56.012302.160310. [DOI] [PubMed] [Google Scholar]
- 43.Biswas S, Mohammad MM, Movileanu L, van den Berg B. Crystal structure of the outer membrane protein OpdK from Pseudomonas aeruginosa. Structure. 2008;16:1027–1035. doi: 10.1016/j.str.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Biswas S, Mohammad MM, Patel DR, Movileanu L, van den Berg B. Structural insight into OprD substrate specificity. Nat. Struct. Mol. Biol. 2007;14:1108–1109. doi: 10.1038/nsmb1304. [DOI] [PubMed] [Google Scholar]
- 45.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 46.Kroetz D, Ahituv N, Burchard E, Guo S, Sali A, Giacomini K. The Pharmacogenomics Center of the University of California, Sam Francisco: at the interface of genomics, biological mechanism and drug therapy. Pharmacogenomics. 2009;10:1569–1576. doi: 10.2217/pgs.09.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlessinger A, Matsson P, Shima JE, Pieper U, Yee SW, Kelly L, Apeltsin L, Stroud RM, Ferrin TE, Giacomini KM, et al. Comparison of human solute carriers. Protein Sci. 2010;19:412–428. doi: 10.1002/pro.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnold K, Kiefer F, Kopp J, Battey JN, Podvinec M, Westbrook JD, Berman HM, Bordoli L, Schwede T. The protein model portal. J. Struct. Func. Genomics. 2009;10:1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, Klein TE. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol. Cell Proteomics. 2005;4:1862–1875. doi: 10.1074/mcp.M500097-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Davis FP, Barkan DT, Eswar N, McKerrow JH, Sali A. Host pathogen protein interactions predicted by comparative modeling. Protein Sci. 2007;16:2585–2596. doi: 10.1110/ps.073228407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suerbaum S, Michetti P. Helicobacter pylori infection. N. Engl. J. Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 53.Mochalkin I, Miller JR, Narasimhan L, Thanabal V, Erdman P, Cox PB, Prasad JV, Lightle S, Huband MD, Stover CK. Discovery of antibacterial biotin carboxylase inhibitors by virtual screening and fragment-based approaches. ACS Chem. Biol. 2009;4:473–483. doi: 10.1021/cb9000102. [DOI] [PubMed] [Google Scholar]
- 54.Marti-Renom MA, Rossi A, Al-Shahrour F, Davis FP, Pieper U, Dopazo J, Sali A. The AnnoLite and AnnoLyze programs for comparative annotation of protein structures. BMC Bioinformatics. 2007;8:S4. doi: 10.1186/1471-2105-8-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orti L, Carbajo R, Pieper U, Eswar N, Maurer S, Rai A, Taylor G, Todd M, Pineda-Lucena A, Sali A, et al. A kernel for open source drug discovery in tropical diseases. PLoS Negl. Trop. Dis. 2009;3:e418. doi: 10.1371/journal.pntd.0000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datta RS, Meacham C, Samad B, Neyer C, Sjolander K. Berkeley PHOG: PhyloFacts orthology group prediction web server. Nucleic Acids Res. 2009;37:W84–W89. doi: 10.1093/nar/gkp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schomburg I, Chang A, Schomburg D. BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 2002;30:47–49. doi: 10.1093/nar/30.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sankararaman S, Kolaczkowski B, Sjolander K. INTREPID: a web server for prediction of functionally important residues by evolutionary analysis. Nucleic Acids Res. 2009;37:W390–W395. doi: 10.1093/nar/gkp339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sankararaman S, Sjolander K. INTREPID—INformation-theoretic TREe traversal for Protein functional site IDentification. Bioinformatics. 2008;24:2445–2452. doi: 10.1093/bioinformatics/btn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kondo H, Shiratsuchi K, Yoshimoto T, Masuda T, Kitazono A, Tsuru D, Anai M, Sekiguchi M, Tanabe T. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc. Natl Acad. Sci. USA. 1991;88:9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waldrop GL, Rayment I, Holden HM. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 64.Mochalkin I, Miller JR, Evdokimov A, Lightle S, Yan C, Stover CK, Waldrop GL. Structural evidence for substrate-induced synergism and half-sites reactivity in biotin carboxylase. Protein Sci. 2008;17:1706–1718. doi: 10.1110/ps.035584.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, et al. Remediation of the protein data bank archive. Nucleic Acids Res. 2008;36:D426–D433. doi: 10.1093/nar/gkm937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kraulis PJ. MOLSCRIPT: A Program to produce both detailed and schematic plorts of protein structures. J. App. Crystallography. 1991;24:946–950. [Google Scholar]
- 67.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Met. Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 68.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver DE, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics research network and knowledge base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 72.Pegg SC, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 73.Schwede T, Sali A, Honig B, Levitt M, Berman H, Jones D, Brenner S, Burley S, Das R, Dokholyan N, et al. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17:151–159. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giglia E. New year, new PubMed. Eur. J. Phys. Rehabil. Med. 2009;45:155–159. [PubMed] [Google Scholar]