Abstract
DroID (http://droidb.org/), the Drosophila Interactions Database, is a comprehensive public resource for Drosophila gene and protein interactions. DroID contains genetic interactions and experimentally detected protein–protein interactions curated from the literature and from external databases, and predicted protein interactions based on experiments in other species. Protein interactions are annotated with experimental details and periodically updated confidence scores. Data in DroID is accessible through user-friendly, intuitive interfaces that allow simple or advanced searches and graphical visualization of interaction networks. DroID has been expanded to include interaction types that enable more complete analyses of the genetic networks that underlie biological processes. In addition to protein–protein and genetic interactions, the database now includes transcription factor–gene and regulatory RNA–gene interactions. In addition, DroID now has more gene expression data that can be used to search and filter interaction networks. Orthologous gene mappings of Drosophila genes to other organisms are also available to facilitate finding interactions based on gene names and identifiers for a number of common model organisms and humans. Improvements have been made to the web and graphical interfaces to help biologists gain a comprehensive view of the interaction networks relevant to the genes and systems that they study.
INTRODUCTION
Networks of interacting genes and gene products mediate most cellular processes. Biologists increasingly appreciate the need to view a gene or protein of interest in the context of its interaction network in order to understand its function. Moreover, studies aimed at understanding how a biological system works require analyses of comprehensive interaction data for all the genes and proteins involved. While this data is available for an increasing number of genes and proteins, the process of finding and integrating it remains a challenge for many biologists. The most extensive interaction data currently available is for protein–protein interactions (PPI). PPI data curated from published literature is available from several large centralized repositories, including BioGRID, IntAct and MINT (1–3). While some PPI data is available in all of these databases, each has a different focus and a set of unique interactions not found in the others. To get a complete set of interactions for any set of proteins, therefore, data from all sources must be integrated, which is a non-trivial task. Additionally, because generic databases include data from many organisms, they frequently lack useful organism-specific information such as phenotypes, expression data and organism-specific vocabularies, and they have no direct means for identifying interactions that are potentially conserved between organisms. DroID, the Drosophila Interactions Database was established to address these problems and to provide a single comprehensive resource for Drosophila PPI (4,5). DroID combines Drosophila PPIs from all available sources as well as predicted PPIs based on experimentally detected PPIs in other organisms. Providing this comprehensive collection in a single database allows users to quickly and easily access all potentially relevant PPIs for a Drosophila protein or group of proteins and eliminates the time consuming and potentially prohibitive effort that would be required to assemble this data from multiple sources. The organism-specific data contained within DroID facilitates efforts to understand the biological significance of individual PPIs and PPI networks.
While protein interactions form the core of many biological pathways, other types of interactions also play crucial roles [Supplementary Figure S1 and (6)]. Many signal transduction pathways, for example, consist of a series of PPI that lead to the activation or inhibition of a transcription factor (TF). TFs in turn bind to and activate or repress the transcription of specific genes. Sequence-specific, DNA-binding TFs help define the set of genes and proteins that are expressed in a particular cell under specific conditions. In order to obtain a complete understanding of many biological pathways, therefore, it would be necessary to integrate TF–gene interaction data with PPI data. Gene expression can also be controlled by regulatory RNAs or non-coding RNAs (ncRNAs). Some ncRNAs regulate the expression of specific genes. For example, microRNAs (miRNAs or miRs) post-transcriptionally regulate specific genes by base pairing with target mRNAs (7). These functional interactions can be referred to as ‘RNA-gene’ interactions, indicating that a particular RNA regulates a particular gene. Recent appreciation of the widespread involvement of RNA–gene interactions in regulating gene expression suggests that most biological processes are likely to involve a combination of PPI, TF–gene and RNA–gene interactions, as illustrated in the example in Supplementary Figure S1. Thus, to fully understand any biological system, data for each of these types of interaction must be combined and ultimately integrated into models for how the system functions. A number of elegant studies have demonstrated the value of integrating different types of interaction data [for recent examples see (8–11)]. However, similar to PPI data, data for other interaction types is spread over many different databases making the processes of collection and integration non-trivial. The new DroID begins to address this problem by collecting and combining PPI and genetic interactions along with TF–gene and RNA–gene interactions into a single frequently updated database where they can be accessed and analyzed together.
A majority of the currently available PPI data has been derived from two-hybrid experiments performed in yeast or from overexpression/pull-down experiments performed in cultured cells. Unfortunately, neither of these assays captures the spatiotemporal variation in the expression of genes in a living organism. Instead, the data is effectively a composite of potential interactions in an organism. In vivo, a gene product may have one set of interaction partners in one cell type and a different set in another or at a different time. In order to obtain a meaningful picture of a biological process then, a researcher must integrate spatiotemporal gene expression patterns with the interaction data. Currently, few databases allow integration of expression and interaction data. The new DroID integrates gene expression data with interaction data, which allows users to assess the potential relevance of interaction data for specific tissues and specific developmental stages.
DATABASE CONTENT
Gene information
DroID currently has interaction data for over 14 000 genes, most of which are protein-coding genes (Table 1). DroID maintains updated and searchable information on all of these genes. Most of the gene information comes from Flybase (12), the authoritative resource for Drosophila gene and genome information. Each gene page of Flybase.org has a link to the relevant gene page in DroID, which takes users to the interaction summary page for the gene (Figure 1). DroID interfaces in turn allow searches of all Flybase gene attributes located in routinely updated DroID tables or via the Flybase Chado database (13). DroID uses Flybase gene identifier numbers (FBgn) as the primary identifiers for genes and their encoded proteins or RNAs. The process of finding interactions of interest generally begins with a search for a gene or set of genes, which once found are then used to search the interaction data. Users can find genes in DroID using gene names or identifiers, or gene attributes such as Gene Ontology terms (14,15), gene class, phenotypes and expression terms. These and other attributes can also be used to filter interaction data or to manipulate the data in an interaction map.
Table 1.
Interactions in DroID
| Data set | Genes | Interactions |
|---|---|---|
| PPI: curagen YTH | 6819 | 20 044 |
| PPI: finley YTH | 3639 | 9069 |
| PPI: hybrigenics YTH | 1274 | 1848 |
| PPI: from other databases | 1104 | 1711 |
| PPI: yeast interologs | 2783 | 76 410 |
| PPI: worm interologs | 1693 | 2996 |
| PPI: human interologs | 4641 | 56 798 |
| GI: genetic interactions | 2906 | 7174 |
| DPI: TF–gene interactions | 139 | 235 |
| RRI: RNA–gene interactions | 9710 | 54 176 |
| Total | 14 204 | 216 593 |
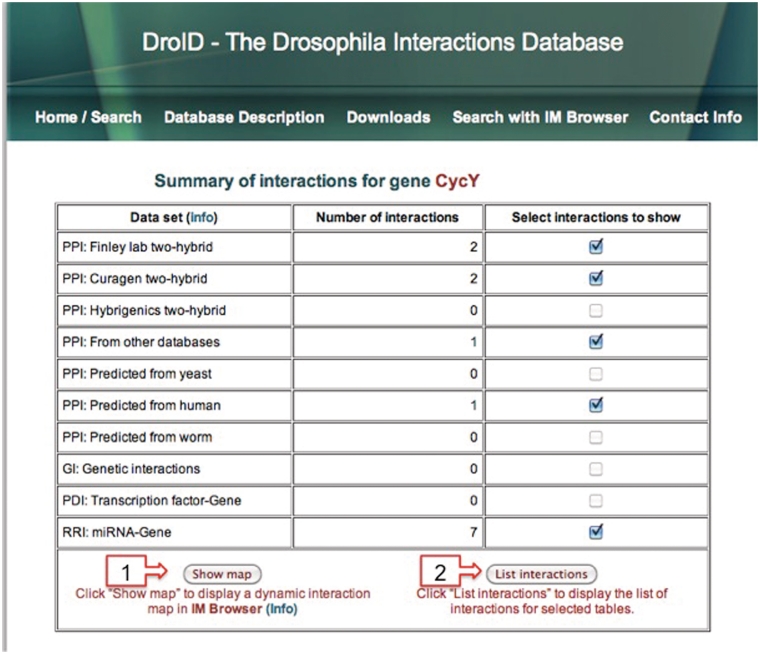
Figure 1.
The DroID Summary of Interactions page. This page summarizes the numbers of interactions in each interaction data set for one or more genes. In this case, the interactions for the Drosophila gene CycY (FBgn0032378) are shown. Users can select the data set that they want to show (all are selected by default) and then choose to view the interaction map in IM Browser (1) or to list the interactions in a web page (2) as in Figure 2. Summary pages for each gene can be generated by inserting the desired FBgn number (#) in the link: www.droidb.org/ppisummary.jsp?gene_ids_for_imbrowser=FBgn#.
A new feature of DroID is gene ortholog information, which should extend the usefulness of the database to beyond the community of Drosophila biologists. Drosophila boasts a particularly large number of mapped gene and protein interactions. Combining this fact with the relatively high level of conservation of genes and pathways between Drosophila and other multi-cellular organisms (including humans), suggests that scientists studying genes and pathways in other organisms could gain insights from the data contained within DroID that may not be available for the organism they are studying. To facilitate the use of DroID by scientists studying other organisms, the new version of DroID contains gene names and identifiers for the Drosophila orthologs of human genes and genes from other model organisms including yeast, worm, zebra fish, frog and mouse. These mappings, which are based on the InParanoid (16) orthology mapping algorithm, act as synonyms for the Drosophila genes thereby expanding the number of different gene names or identifiers that can be used to search for Drosophila genes. For example, users familiar only with the human or yeast name of a gene could readily search DroID for interactions involving the Drosophila orthologs.
PPIs
DroID gathers all of the PPI data from large-scale studies and from external databases into a single resource. The largest amount of PPI data for Drosophila comes from large-scale yeast two-hybrid studies, which together have combined to identify more than 24 000 interactions (17–19). DroID also includes all of the Drosophila PPI data from the literature-curated multi-organism databases, BioGRID, IntAct and MINT (1–3); since each of these databases has a unique set of interactions not found in the others (Supplementary Figure S2), DroID is currently the only place where all of the data can be found and analyzed together. A significant addition to the new DroID is the inclusion of unpublished Drosophila PPIs as they are being generated in an ongoing high-throughput yeast two-hybrid screen (www.proteome.wayne.edu). Currently there are more than 6000 of these interactions available in DroID. To supplement the experimentally determined PPI, DroID also contains predicted Drosophila PPI that are based on the large amount of PPI data that has been generated for other organisms. We obtain experimental PPI data from BioGRID, IntAct and MINT for Saccharomyces cerevisiae and Caenorhabditis elegans, and from the same databases plus Reactome (20) and HPRD (21) for human, and we map them onto the orthologous Drosophila proteins to generate predicted conserved interactions, or interologs. Again, each of the source databases contributes a unique set of interologs to DroID (Supplementary Figure S3). Inclusion of these potentially conserved interactions expands the available PPIs in DroID by more than 160 000 interactions in the most recent update. Collecting PPIs from all of the different available sources allows DroID to function as a complete resource of Drosophila PPIs for the research community.
To keep pace with the almost continuous stream of newly generated PPI and other interaction data, DroID is regularly updated. New to DroID is a streamlined protocol for updating the gene and interaction data. The update protocol combines automatic collection of data from external databases with manual curation of the collected data sets and high-throughput data. The data is combined and checked for consistency, interologs are generated and identifiers are updated to the latest annotation of the genome available in Flybase. A complete update of the database can now be completed in less than one day, which allows for an improved update schedule of approximately every month.
New interaction types: TF–gene and RNA–gene
The new DroID now includes tables for TF–gene interactions that can be searched separately or integrated with the PPI and genetic interaction tables. The demonstration that a TF can bind to a cis-regulatory module (CRM) for a gene suggests that the TF may regulate the gene, though experimental confirmation that the TF directly regulates the gene in vivo is often not available. Despite this uncertainty, TF–gene data can provide insights into potential transcription regulatory networks. Currently, the TF–gene interaction table includes data from REDfly (22), a continuously updated database of interactions curated from published literature. The high-quality data in REDfly currently includes only interactions demonstrated experimentally by DNaseI footprinting assays along with CRMs that have been verified by experimental methods such as reporter gene assays. Consistent with the philosophy of DroID, we have included all information that is available for these interactions including links to the original source publications and to the source REDfly pages, the CRMs to which the TF binds and available expression data using controlled vocabularies (CVs) (see below). All of this data can be searched and used to find or filter interaction data sets.
In contrast to the burgeoning TF–gene interaction data, currently there are very few experimentally verified RNA–gene interactions for Drosophila. Most sources of RNA–gene interactions, therefore, primarily include interactions predicted using base pair complementarity between miRNAs (miR) and their putative targets (23). In Drosophila, over 140 miRs have been identified thus far and more are likely to follow (24). Each miR has from a few to hundreds of predicted targets, suggesting that expression of a particular miR could profoundly influence the genetic networks that are active in any particular cell or developmental time point. The new DroID contains predicted miR-target gene data for 146 miRs belonging to 122 miR families from the TargetScanFly (25) database. The miR identifiers have been mapped to Flybase gene identifiers using information available at Flybase and MirBase (26). Again, DroID includes all information available for these interactions including information on whether the prediction is for conserved miR families or non-conserved miR families, the target site conservation and the original prediction database.
Gene expression data
DroID includes three different types of data that enable users to search and filter interactions based on gene expression patterns. First, DroID includes correlation values that indicate how similar the expression patterns are for interacting genes. Pairs of genes that are frequently upregulated or downregulated together across many different gene expression experiments, for example, have a higher expression correlation value (e.g. closer to 1) than genes that are not frequently expressed together. DroID computes the correlation values based on the extensive genome-wide expression data available in the NCBI Gene Expression Omnibus (GEO) (27). The correlation values can be used to filter interaction data in order to focus on networks of genes that are frequently expressed together and therefore more likely to function together. These correlation values are also useful for a variety of other analyses and are available for download at the DroID website. Second, DroID includes data derived from microarray studies that examined RNA expression at different developmental stages (28,29). This data can be used to focus on interactions among genes that are expressed at user-defined stages and levels. The third type of expression data now in DroID is RNA and polypeptide localization data curated by Flybase. The data is from published literature and from ongoing large-scale projects (28), includes both RNA and polypeptide localization data where available, and is annotated using controlled vocabularies. DroID downloads and processes this information and stores it in the gene attributes table. Along with the already existing expression filters in DroID, this new data will help biologists to explore and understand the dynamic aspects of interaction networks.
ACCESS INTERFACES
DroID data can be browsed and retrieved via three easy-to-use interfaces, including a web site (www.droidb.org) implemented with Java EE technology, a graphical web application called Interaction Map (IM) Browser (4) implemented as a Java applet, and a DroID plug-in for the network visualization and analysis program, Cytoscape (30). Each interface allows users to search for one or more Drosophila genes based on a variety of gene attributes. Once a gene or set of genes is found, users can select to search for interactions involving it in specific interaction data sets. IM Browser and the DroID Cytoscape plug-in also allow more sophisticated searches based on attributes of specific interaction data sets. All three interfaces also have features for filtering interactions based on PPI confidence scores, gene expression data and any gene or interaction attribute. The main features of the interfaces are outlined below. Tutorials and help pages are available at the DroID website (www.droidb.org).
Web interface
The web pages offer rapid access to DroID data and include a number of new or improved search and filter tools (Figures 1 and 2). The DroID home page (www.droidb.org) is also the main search page and allows a user to search for fly genes using one or more gene identifiers; e.g. gene name, gene symbol, synonym, FBgn identifier. Gene searches can also be based on gene ontology, gene class, protein domains and gene expression patterns. Researchers studying other organisms can make use of the large amount of interaction data in DroID by selecting their organism of interest and typing the organism-specific gene name or identifier in the search box. Genes that satisfy the search criteria will be displayed in a new page that allows the user to choose which of the displayed genes is to be used for their query. The user is also presented with a choice of which interaction data sets they want queried. DroID will then search the data sets using the selected genes and display the results in a results page (Figure 2). The results page lists the found interactions, the data sets that they were found in, and in the case of PPIs a confidence score. From the results page users can choose to view additional gene information via an expandable link that reveals gene ontology, expression patterns and links to relevant databases such as FlyBase. Each gene also has a link that will display a summary of the interactions in DroID for that gene. Users can also expand the ‘Found in’ link to view experimental details relevant to each data set in which the interaction was found, including a list of the databases that originally curated the data and links to the PubMed (www.ncbi.nlm.nih.gov/pubmed) citations for literature that originally described the interaction. Filters can be applied to the list of interactions based on confidence scores, expression correlation values, genes expression levels in microarray studies or stage and tissue gene expression patterns (Figure 2 and Supplementary Figure S4). Finally, information from the results page can be downloaded for offline analysis.
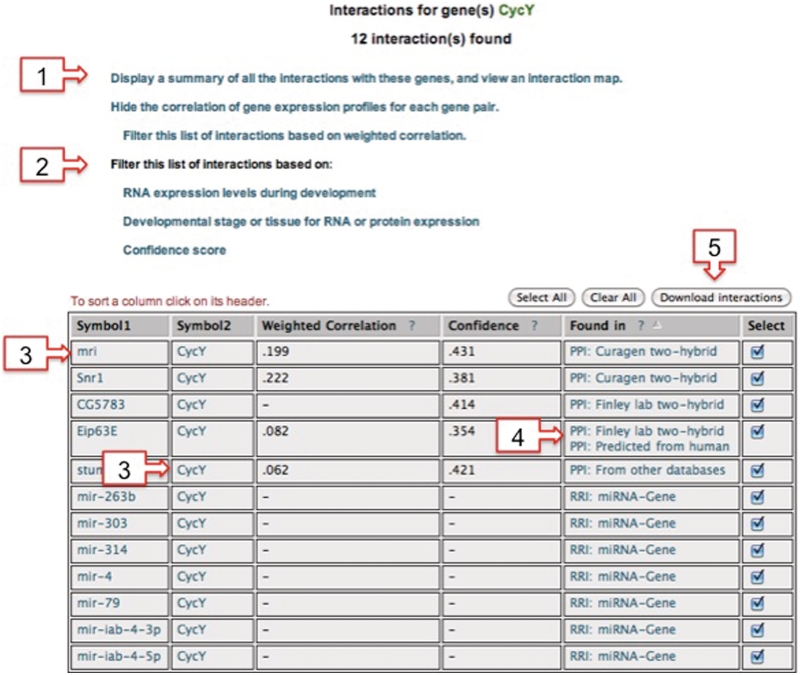
Figure 2.
The Interaction List results page. This results page can be reached from the Summary of Interactions page (Figure 1) or from the Home/Search page after first searching for the gene of interest and selecting the desired interaction data set(s) to search. The page includes a number of links: (1) a link to the Summary of Interactions page; (2) links that open windows to enable filtering the interaction list based on expression correlation values, RNA expression levels, RNA or protein expression patterns or PPI confidence scores (see Supplementary Figure S4 for examples); (3) detailed information and links to external databases are available for each gene by clicking on the gene symbol; (4) detailed information and links are also available by clicking on the data sets listed in the ‘Found in’ column; (5) clicking on the download button will download the data from the table; users are given the option to also download other details.
Network visualization tools
To view and manipulate a graphical interaction map of DroID data, users can either download the Cytoscape DroID plugin or use the interaction map program, IM Browser, which has undergone a number of recent improvements. The IM Browser interface allows users to perform advanced queries of the DroID database and to visualize the results in a dynamic graphing environment (Figure 3). Users can input a single gene or upload a list of genes into the IM Browser query form or they can search for genes through a connection between IM Browser and the Flybase database. Once query gene(s) have been selected users then choose which of the DroID interaction data sets they want queried. Users can also search with a list of interactions, essentially asking whether any of the interactions are in DroID, or search for networks that connect any pair of genes. In the latter case, the search will find the sub-network that includes the shortest path(s) between the two genes. Additionally, users can upload their own interactions from a local file, enabling them to search, view and filter their data and integrate it with DroID data. Visualizing the results of a query as an interaction map allows the user to recognize patterns and relationships that are not easily appreciated by viewing a table or list of interactions. Nodes (genes, proteins or RNAs) and edges (interactions) are clickable to obtain additional information including links to external databases. The interaction map layout can be manipulated and the colors and shapes of nodes or edges can be changed based on user-defined schemes or based on gene and interaction attributes in DroID or Flybase. IM Browser makes essentially all of the data in DroID available for searches in order to add new interactions to a map or to filter existing maps. At the completion of an IM Browser session users can download an image of the interaction map or a table that includes user-chosen node and edge attributes. Options are also available for downloading an XML file that includes all of the session’s queries so that the same queries can be run again in the future, for example, on updated versions of DroID data.
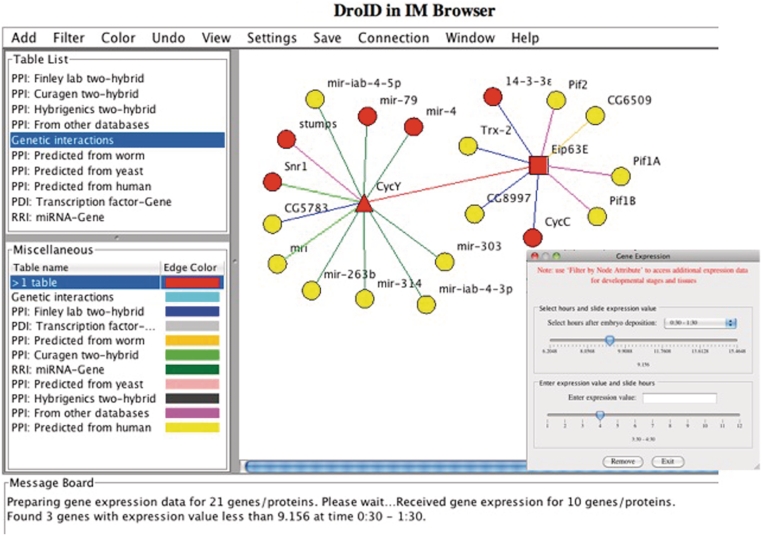
Figure 3.
Exploring DroID networks with IM Browser. An interaction map centered on the Drosophila proteins CycY and Eip63E is shown. Interaction lines are colored according to the data source. Nodes were colored red by searching the map for genes that are expressed at the embryonic stage (under the Color menu). The Add, Filter and Color options allow users to add, remove or color interactions and nodes based on all of the gene and interaction data in DroID. One filter window is also shown; this filter allows users to slide a scale to either remove or add back nodes based on their embryonic expression levels. Other filters allow access to the RNA and protein expression patterns. Right clicking on any node, edge or interaction data set presents more detailed information including additional options for searching, filtering and coloring the map. The layout can be manipulated manually or by using preset layouts under the Settings menu. The Save menu presents options for saving the data as a figure or table, or as a query file that can be run again to reproduce the map in the future.
FUTURE DEVELOPMENT
Several further improvements to DroID and its interfaces are ongoing or planned. First, we intend to expand our support for the Protein Standards Initiative Molecular Interaction (PSI-MI) format (31). DroID currently includes data fields compatible with PSI-MI, including the PSI-MI ontology terms and identifiers when they are available for an interaction. In addition, DroID maintains the proposed minimal information required to report a molecular interaction (32), when that information is available from other databases or the literature. Users, therefore, can download DroID data and convert it to the PSI-MI format using one of the publically available tools for that purpose (e.g. see www.psidev.info). Future releases of DroID will include options to download the data directly in the PSI-MI format to facilitate analysis of the data with other programs. Second, future updates of DroID will expand the TF–gene data to include data from genome-wide studies such as the ModEncode project (www.modencode.org) aimed at identifying in vivo binding sites for TFs (33,34). As with PPI data, DroID will strive to represent the uncertainty associated with this data; e.g. although the finding that a TF binds to a gene under specific conditions does not necessarily mean that the TF regulates the gene, it does provide an important first clue. Third, we will also expand the sources from which RNA–gene interactions are obtained. Initially, we will present the results from different miR target prediction algorithms in addition to TargetScan, such as MiRanda (35), PicTar (36) and MinoTar (37). We will also strive to include information on experimental verification of miR and their targets as it becomes available, for example, from existing curation efforts [see, for example, TarBase; (38)]. Fourth, the DroID database and interfaces are capable of accommodating additional functional information about interactions and we hope to include this data as it becomes available. For example, a key piece of information that is currently missing from much of the TF–gene and RNA–gene data is whether or not an interaction represents an activating or repressing relationship. We expect this information to be increasingly available and will incorporate it into DroID when possible. Finally, we plan to continue to update the web and graphical interfaces to DroID. User feedback will continue to inspire and drive future improvements.
CONCLUSIONS
DroID was originally intended to serve as a centralized database for all Drosophila PPIs. Appreciation for the importance of additional interaction types and the necessity to integrate these interactions with PPIs has driven the evolution of the database. The new DroID serves not only as a comprehensive resource for Drosophila PPI data, but also as a tool for combining PPI data with TF–gene, RNA–gene and gene–gene data to construct and explore more complete biological networks. The combination of the multiple interaction types with the spatiotemporal gene expression data in DroID allows users to obtain snapshots of interaction networks that are relevant to specific developmental times and tissues. As the amount and diversity of interaction data that is available continues to increase, DroID and similar databases that allow users to access and interpret comprehensive gene and protein interaction data will be essential tools for the study of biological pathways and networks.
SUPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (grant numbers HG001536 and P41RR18327 to R.L.F.). Funding for open access charge: NIH grant (HG001536) and Wayne State University funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank members of the Finley laboratory and the Drosophila research community for providing valuable feedback on the continued development of DroID. We also thank members of the Finley laboratory for unpublished data and the public databases referenced in the text for their outstanding curation efforts. We are particularly grateful to Dr Mark Halfon for providing RedFly data and Dr David Bartel for providing TargetScanFly data.
REFERENCES
- 1.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacifico S, Liu G, Guest S, Parrish JR, Fotouhi F, Finley RL., Jr A database and tool, IM Browser, for exploring and integrating emerging gene and protein interaction data for Drosophila. BMC Bioinformatics. 2006;7:195. doi: 10.1186/1471-2105-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, Pacifico S, Liu G, Finley RL., Jr DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanyon CA, Finley RL., Jr Progress and potential of Drosophila protein interaction maps. Pharmacogenomics. 2000;1:417–431. doi: 10.1517/14622416.1.4.417. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, Bumgarner RE, Schadt EE. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexeyenko A, Sonnhammer EL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107–1116. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat. Genet. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 14.Consortium GO. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 19.Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, Zhang H, Zhong J, Finley RL., Jr A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasad TS, Kandasamy K, Pandey A. Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol. Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- 22.Halfon MS, Gallo SM, Bergman CM. REDfly 2.0: an integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila. Nucleic Acids Res. 2008;36:D594–D598. doi: 10.1093/nar/gkm876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1209s29. Chapter 12, Unit 12 1911–10. [DOI] [PubMed] [Google Scholar]
- 27.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 30.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stumpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 33.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonn S, Furlong EE. cis-Regulatory networks during development: a view of Drosophila. Curr. Opin Genet. Dev. 2008;18:513–520. doi: 10.1016/j.gde.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3'UTRs. Proc. Natl Acad. Sci. USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]