Abstract
miRGator is an integrated database of microRNA (miRNA)-associated gene expression, target prediction, disease association and genomic annotation, which aims to facilitate functional investigation of miRNAs. The recent version of miRGator v2.0 contains information about (i) human miRNA expression profiles under various experimental conditions, (ii) paired expression profiles of both mRNAs and miRNAs, (iii) gene expression profiles under miRNA-perturbation (e.g. miRNA knockout and overexpression), (iv) known/predicted miRNA targets and (v) miRNA-disease associations. In total, >8000 miRNA expression profiles, ∼300 miRNA-perturbed gene expression profiles and ∼2000 mRNA expression profiles are compiled with manually curated annotations on disease, tissue type and perturbation. By integrating these data sets, a series of novel associations (miRNA–miRNA, miRNA–disease and miRNA–target) is extracted via shared features. For example, differentially expressed genes (DEGs) after miRNA knockout were systematically compared against miRNA targets. Likewise, differentially expressed miRNAs (DEmiRs) were compared with disease-associated miRNAs. Additionally, miRNA expression and disease-phenotype profiles revealed miRNA pairs whose expression was regulated in parallel in various experimental and disease conditions. Complex associations are readily accessible using an interactive network visualization interface. The miRGator v2.0 serves as a reference database to investigate miRNA expression and function (http://miRGator.kobic.re.kr).
INTRODUCTION
MicroRNAs (miRNAs) are important post-transcriptional regulators that are associated with various cell functions and human diseases. MiRNAs bind to complementary sequences in the 3′ UTRs of target mRNAs resulting in negative regulation of gene expression (1,2). Significant efforts have been made to study the function of miRNAs and their target genes. However, the function of most miRNAs are still elusive and under active investigation. A number of miRNA-related databases have been developed. There are many miRNA target prediction algorithms available including TargetScan (3), PITA (4), miRanda (5), PicTar (6), miBridge (7) and many more. PhenomiR (8) and miR2Disease (9) provide information on disease-associated miRNAs from literature curation. FAME (10) uses a target prediction method to infer biological processes affected by human miRNAs. MMIA (11) and MAGIA (12) provide tools for gene (or miRNA) set analysis and target prediction using miRNA–miRNA expression profiles.
MiRGator is an integrated database of miRNA-associated gene expression, target prediction, disease association and genomic annotation, which aims to facilitate functional investigation of miRNAs. In this update version, we integrate a variety of publicly available data sets related to miRNA biology. Overall, more than 10 000 miRNA/mRNA expression profiles are compiled and organized by manual curation according to miRNA, tissue type, disease and perturbation. MiRGator contains information about (i) human miRNA expression profiles under various experimental conditions, (ii) paired expression profiles of both mRNAs and miRNAs from the same sample, (iii) gene expression profiles under miRNA-perturbation (e.g. miRNA knockout and overexpression), (iv) known/predicted miRNA targets and (v) miRNA-disease associations. In addition, a number of new features have been added to the previous version, such as association analysis among miRNAs, diseases and perturbations and an integrated network viewer. Here, an overview of the miRGator system, the collection and processing of data sets, and important features are briefly described. A more detailed usage and documentation are provided on-line (http://miRGator.kobic.re.kr).
SYSTEM OVERVIEW
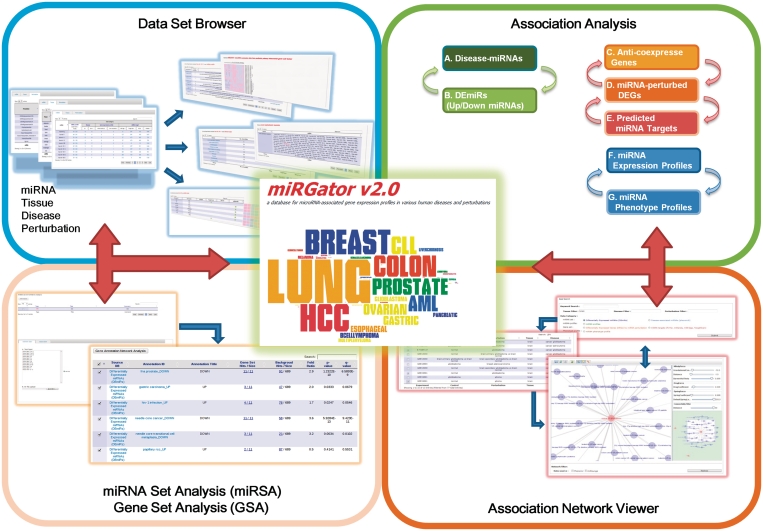
The miRGator v2.0 consists of three main modules: the data set browser, the association analysis and the gene set analysis (GSA)/miRNA set analysis (miRSA) modules. The overall system of miRGator v2.0 is shown in Figure 1. Each of these three modules has its distinct function but they are tightly integrated to facilitate functional investigation of miRNAs. Particularly, the expression data sets are manually annotated according to miRNA, target, tissue type, disease and perturbation, so that the user can easily focus on the subject of interest.
Figure 1.
System overview of miRGator v2.0. There are three main modules. The data set browser allows access to all the data sets by miRNA, tissue, disease and perturbation. Within the association analysis module, miRNAs, diseases and perturbations can be associated by GSA, miRSA and miRPA. The resulting association network is visualized by an integrated association network viewer. In the GSA/miRSA module, the user may enter a list of miRNAs/genes and perform GSA/miRSA against miRNA sets or gene sets in the miRGator system.
The data set browser provides diverse access routes to our comprehensive miRNA-related data sets by miRNA, tissue type, etc. The data are hierarchically organized in several tables with increasing details from top to bottom, e.g. from the full list of miRNAs to the actual expression values of each miRNA. The association analysis connects different miRNAs, diseases and perturbations by the overlap among miRNA/gene sets and by the similarity between miRNA expression/phenotype profiles. The association analysis module is also equipped with an integrated association network viewer for convenient exploration of association networks. Finally, the GSA/miRSA module takes user-defined gene sets or miRNA sets and performs GSA or miRSA against various types of gene annotations [e.g. gene ontology (GO, 13), KEGG (14)] as well as the miRGator data sets.
DATA SOURCES AND PROCESSING
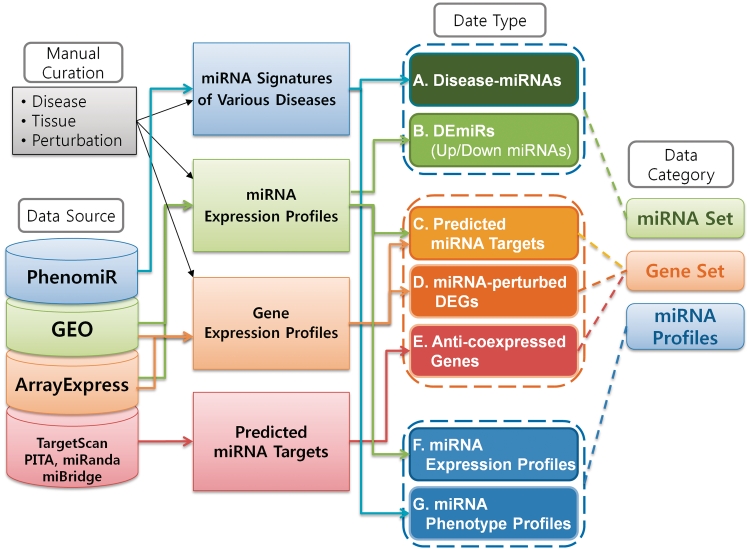
The core data set of miRGator v2.0 consists of seven different data types collected from various sources such as gene expression, miRNA-disease association and four major miRNA-target databases (TargetScan, PITA, miRanda and miBridge). The seven data types belong to one of the three broad categories―miRNA set, gene set and miRNA profiles. Where appropriate, these data sets were manually curated according to tissue, disease and perturbation (e.g. chemical treatment). The overall procedure of data collection and processing is shown in Figure 2.
Figure 2.
The overall procedure of data collection and processing.
The miRNA set category is basically a list of miRNAs. This category consists of known disease-miRNA associations from PhenomiR (8) and differentially expressed miRNAs (DEmiRs) from the gene expression omnibus (GEO) (15) and ArrayExpress (16). For DEmiRs, we collected miRNA expression data for various cancers and perturbations. In total, 154 miRNA expression data sets (8431 profiles) were downloaded from the GEO and ArrayExpress. The miRNA expression profiles were quantile normalized using LIMMA package (17). The miRNA profiles were hand-curated, resulting in the annotations of 26 human tissue types, 56 diseases and 83 perturbations.
The gene set category contains information for miRNA targets such as predicted miRNA targets, anti-coexpressed miRNA-gene pairs and miRNA-perturbed differentially expressed genes (DEGs) (i.e. the up-regulated genes under miRNA-knockout conditions and down-regulated genes under miRNA-overexpression conditions). The predicted miRNA targets were collected from TargetScan, PITA, miRanda and miBridge. The miRNA-perturbed gene expression profiles were obtained from GEO. We compiled 68 data sets of 613 gene expression profiles, where one or more miRNAs were knocked out/down or overexpressed. After filtering out inappropriate sets, the remaining 63 sets contain 595 profiles annotated with 88 miRNA perturbations (90 different miRNAs), 15 tissues and 18 diseases. For anti-coexpressed gene sets, we collected 22 data sets from GEO, where both miRNA and mRNA expression profiles were measured for the same sample. The total number of the paired miRNA and mRNA profiles was 1265 and 1568, respectively. Pearson correlation between miRNA and mRNA expression values was calculated for each of the 22 data sets. Accordingly, a miRNA–mRNA pair may have up to 22 correlation values. We compiled anti-coexpressed miRNA–mRNA pairs, which show significant anti-correlation in at least two or more cases among the 22 data sets at P-value cut off <0.001. For miRNAs with more than 500 anti-correlated mRNAs above cut-off, we took only the top 500 mRNAs ranked by the sum of –log (P-value).
The last category is miRNA-related profile, which consists of miRNA expression profiles and phenotypic profiles. The miRNA expression profiles came from the same data used in the DEmiR set. Phenotypic profiles were obtained from the PhenomiR data describing up/down regulation of miRNAs in various diseases. Additionally, various types of gene annotation information are integrated including GO and biological pathways (KEGG, BioCarta). The details of data source, type and statistics are shown in Table 1 and on-line.
Table 1.
Summary statistics of data sets in miRGator v2.0
| Data category | Data type | Description | Experiments (Profiles) | miRNAs | mRNAs | Diseases | Tissues | Perturbations | Data source |
|---|---|---|---|---|---|---|---|---|---|
| miRNA set | Disease- miRNAs | Disease associated miRNAs | – | 354 (up) 340 (down) | – | 59 | 25 | – | PhenomiR |
| DEmiRs | DEmiRs under diseases/perturbations | 146 (8013) | 689 | – | 47 | 21 | 128 | GEO ArrayExpress | |
| Gene set | miRNA targets | Predicted miRNA target genes | – | 700 | 19 069 | – | – | – | miBridge, TargetScan, PITA, miRanda |
| miRNA- perturbed DEGs | miRNA knockout | 5 (92) | – | 15 708 | 4 | 4 | 16 | GEO | |
| miRNA overexpression | 20 (205) | – | 20 887 | 12 | 12 | 27 | GEO | ||
| Coexpressed genes | Positive and negative coexpression between miRNA and mRNA from paired expression profiles | 22 (2538) | 685 | 27 830 | 11 | 13 | – | GEO | |
| miRNA profiles | Expression profiles | miRNA expression profiles under various conditions | 146 (8013) | 689 | – | 47 | 21 | 128 | GEO ArrayExpress |
| Phenotype profiles | Profiles of disease associations for each miRNA | – | 354 (up) 340 (down) | – | 59 | 25 | – | PhenomiR |
DATA SET BROWSER
From the data set browser, the user can access all the data sets complied in miRGator v2.0, which are organized according to miRNA, target, tissue type, disease and perturbation. Accordingly, the user may access the data set of interest using various annotation categories of choice. Several tables are sequentially displayed from top to bottom with increasing detail as the user chooses an item of interest. For example, the user can start from the list of all miRNAs, click to show the list of miRNA profiles containing the selected miRNA, and then choose a specific miRNA experiment, where the actual expression values are displayed as a heatmap. MiRNA–disease association, and tissue-specific or disease related expression data can be accessed in a similar manner.
Particularly, miRGator v2.0 is useful to obtain integrated information on miRNA target reliability. The miRNA–target relationship may be supported by multiple, independent evidences such as target prediction, up-regulation on miRNA knockout, down-regulation on miRNA overexpression and miRNA–mRNA anti-coexpression from paired expression experiments. These data supporting the miRNA–target relation are shown side-by-side simultaneously. For example, RAC1 (Entrez Gene ID: 5879) is down-regulated by overexpression of hsa-mir-155 in the GSE14477 data set. RAC1 is predicted as a target by two methods (PITA and miRanda) and also shows a strong negative correlation in two out of the 22 miRNA–mRNA paired expression data sets. This strongly suggests that RAC1 is a genuine target of hsa-mir-155, which needs further experimental validation.
ASSOCIATION ANALYSIS AND ASSOCIATION NETWORK VIEWER
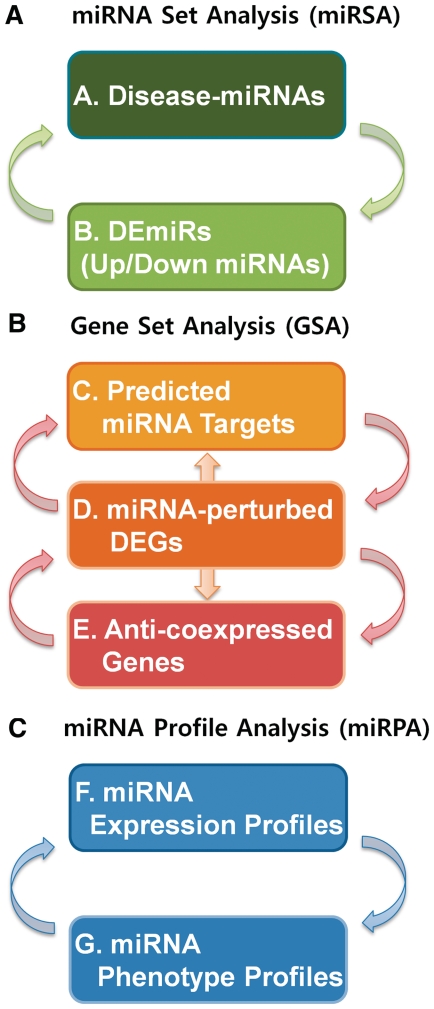
The data sets in miRGator belong to one of the three categories of gene set, miRNA set and miRNA-related profile. Within the data types of the same category, association relationships are established by the overlap among the gene/miRNA sets or by the similarity of profiles (Figure 3). For example, miRNA-perturbed DEGs can be linked to miRNA targets by GSA. Similarly, differentially expressed miRNAs (DEmiRs) can be associated to disease-related miRNAs. In miRNA profile analysis (miRPA), a pair of miRNAs may be functionally related if their expression is regulated in parallel in various experimental and disease conditions as is the case with a pair of genes. Likewise, a pair of miRNAs with similar phenotypic or disease-related patterns is likely to be functionally associated. The resulting associations can be visualized by the association network viewer (bottom right in Figure 1). The network viewer is equipped with useful features such as save network, export image, highlighting nodes interactively, group nodes by disease, etc. and a detailed description is provided in the on-line documentation.
Figure 3.
The three different types of associations in the association analysis module. Depending on the data category, (A) miRSA, (B) GSA and (C) miRPA.
miRSA AND GSA
The GSA/miRSA module allows the user to enter a list of genes and perform GSA/miRSA for statistical enrichment. Association between two sets of miRNAs is also possible in a similar manner to GSA, which we call miRSA. Most representative gene IDs are cross-referenced and automatically converted to the miRBase ID or NCBI entrez gene IDs. For miRSA, we use our own compilation of miRNA sets (i.e. disease-associated miRNAs and DEmiRs; data type A and B in Figure 2) in addition to the PhenomiR database. The GSA is applied for the predicted miRNA targets (data type C), miRNA-perturbed DEGs (data type D), anti-coexpressed genes (data type E). Additional categories include the GO, disease gene [Online Mendelian Inheritance in Man (OMIM, 18) and genetic association database (GAD, 19)], and pathway (KEGG and BioCarta) analyses. The user may enter a list of genes or miRNAs to check any relevance to miRNA biology or existing annotations. On log-in, the user-defined gene/miRNA sets may be saved on the system, which can be retrieved later for further analysis.
CONCLUSION
The miRGator v2.0 integrates human miRNA-related expression patterns under various disease and perturbation conditions. The system is designed to facilitate functional investigation of miRNAs by integrating various evidences of miRNA targets and by revealing non-canonical associations among miRNAs, diseases, perturbations and user-defined gene/miRNA sets.
FUNDING
GIST Systems Biology Infrastructure Establishment Grant (2010) through Ewha Research Center for Systems Biology (ERCSB); Biogreen 21 Program of the Korean Rural Development Administration (20070401034010); National Core Research Center (NCRC) program (R15-2006-020) of the KOSEF funded by the MEST. Funding for open access charge: GIST Systems Biology Infrastructure Establishment Grant (2010) through Ewha Research Center for Systems Biology (ERCSB).
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 5.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, Dunger I, Fobo G, Frishman G, Montrone C, Theis FJ. PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes. Genome Biol. 2010;11:R6. doi: 10.1186/gb-2010-11-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulitsky I, Laurent LC, Shamir R. Towards computational prediction of microRNA function and activity. Nucleic Acids Res. 2010;38:e160. doi: 10.1093/nar/gkq570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–W362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sales G, Coppe A, Bisognin A, Biasiolo M, Bortoluzzi S, Romualdi C. MAGIA, a web-based tool for miRNA and genes integrated analysis. Nucleic Acids Res. 38(Suppl.):W352–W359. doi: 10.1093/nar/gkq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update––from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 18.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nature Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]