Abstract
The polyglutamine diseases are caused in part by a gain-of-function mechanism of neuronal toxicity involving protein conformational changes that result in the formation and deposition of β-sheet rich aggregates. Recent evidence suggests that the misfolding mechanism is context-dependent, and that properties of the host protein, including the domain architecture and location of the repeat tract, can modulate aggregation. In order to allow the bioinformatic investigation of the context of polyglutamines, we have constructed a database, PolyQ (http://pxgrid.med.monash.edu.au/polyq). We have collected the sequences of all human proteins containing runs of seven or more glutamine residues and annotated their sequences with domain information. PolyQ can be interrogated such that the sequence context of polyglutamine repeats in disease and non-disease associated proteins can be investigated.
INTRODUCTION
Polyglutamine (PolyQ) repeats are implicated in several neurodegenerative diseases, including Huntington’s disease and several spinocerebellar ataxia’s. It is commonly thought that a toxic gain-of-function mechanism is triggered by the presence of a polyQ tract, involving a conformational change within the protein and the formation and deposition of β-sheet rich amyloid-like fibrils (1–3).
The length of the polyQ repeat is critical to pathogenesis; however, there is evidence that other protein factors, including the location, type and number of flanking domains can modulate pathogenesis (4–10). Although there are many human polyQ-containing proteins (11), only nine polyQ-containing proteins are implicated in pathogenesis, and the precise repeat threshold to pathogenesis varies within the disease subset, for example, a 37 glutamine repeat is sufficient to lead to Huntington's disease, while SCA3 results only when the polyQ repeat expands to 45 or greater (12–14).
Many other human, non-disease related proteins contain polyQ repeats, which are intrinsically prone to expansion at the genetic level (11,15,16). In fact, a 40 glutamine repeat is the normal allele present in forkhead box P2 transcription factor; a protein that has not been found to be associated with a polyQ disease (17,18). This evidence has led to the hypothesis that protein characteristics modulate the propensity of polyQ-containing proteins to aggregate and cause disease. To investigate the variable characteristics of polyQ proteins we have performed a bioinformatics investigation of the protein context of polyglutamine repeats, and constructed a web-accessible database of all human proteins containing a polyQ repeat greater than seven glutamines in length, termed ‘PolyQ’. The PolyQ database provides a tool to compare the polyQ repeat location, the occurrence/type of domains and the number of domain repeats present across disease and non-disease proteins.
PolyQ DESCRIPTION AND USE
PolyQ was created using open-source MySQL relational database server software, version 5.0.82 (http://www.mysql.com), running on an Apple 8-core 3.0 GHz Xeon/OS X Server (version 10.5.8). The database consists of three tables. A web-based query interface to the database was developed using the PHP5 programming language, hosted via Apache 2.2.14. The user interface was developed with the utilisation of the JQuery Javascript library and JQuery widgets. Charts and graphs are constructed on the fly using the Google Visualization API.
The PolyQ database was populated by extracting all human sequences from the NCBI non-redundant (NR) database that contained at least seven consecutive glutamine residues. We then performed a Pfam (19) domain search to find protein domains within this subset of sequences. The NCBI NR contains many versions of the same protein, which created bias in the statistical analysis of PolyQ location data. We simplified the analysis by indentifying protein variants/isoforms and using only the longest protein isoforms (which we termed ‘master sequences’), therefore eliminating splice variants/protein fragments. Multiple variants/isoforms of each protein were crudely identified by comparing the protein sequence following the PolyQ chains. The original sequences were then subjected to the BLASTClust (20), FORCE (21), MCL (22) and HomoClust algorithms (23), and the variants/isoforms were adjusted as necessary. The crude identification used the 10 amino acids immediately after the PolyQ chain as a ‘search string’; any sequence that had the 10 amino acids immediately following its own polyQ chain was presumed to have homology with that sequence. The homology groups were confirmed by analyzing the data using the above algorithms. This yielded a total of 128 master sequences, from an original data set of >700 polyQ-containing human protein sequences.
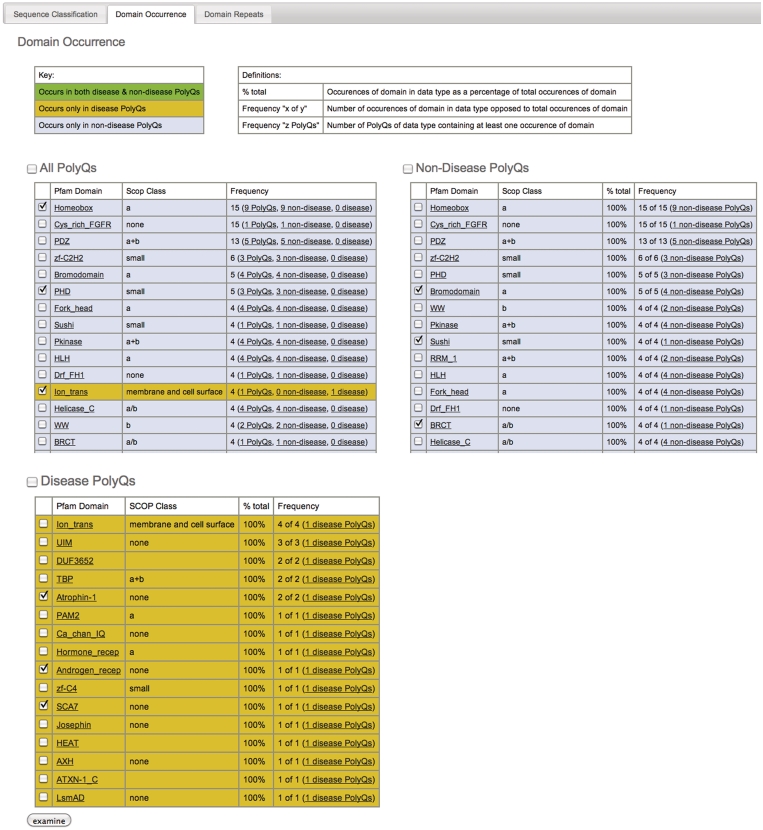
The database can be searched according to protein name, Pfam domain or sequence. The results of a typical search, shown in Figure 1A, show both a graphical summary (Figure 1A, top) and textual details (Figure 1A, bottom) according to sequence classification (see below). The graphical summary shows pie chart and bar chart representations of the results according to sequence classification (Figure 1A, top), Pfam domain occurrence (Figure 1B) and Pfam domain repetition (Figure 1C). Retrieved database entries are listed in table format with one row per protein, and three columns containing protein name (with links to the GenBank entry), Pfam domains, and protein sequence (with the polyQ region annotated), respectively. Homologs in the database can be included or excluded from the search. From this view, the domain and sequence context of the polyQ sequence can be identified and further interrogated. To aid analysis specific entries can be selected from the results (using the ‘examine’ button) and grouped together.
Figure 1.
(A) Typical results of a simple search (blank in this instance), showing graphical breakdown according to sequence classification; Results shown graphically according to domain occurrence (B) and domain repeats (C) using the tabs at top of page [as seen in (A)].
Sequence classification
The data are sorted and annotated according to the following sequence classifications: N-Terminal PolyQs—sequences where the first polyQ chain appears before all Pfam domains; C-Terminal PolyQs—sequences where the last polyQ chain appears after all Pfam domains; Interdomain PolyQs—sequences where the polyQ chains appear between the first Pfam domain and the last Pfam domain; Mid Domain PolyQs—sequences in which the polyQ chain appears in the middle of a Pfam domain, or overlaps a Pfam domain; No Significant Domain PolyQs—sequences that do not contain any significant Pfam domains; Unclassified PolyQs—sequences that did not fit into any of the above classifications. Each group is readily accessed using the tabs in the web page (Figure 1A). We have also further reduced the redundancy in the data by clustering sequence homologs, and have also tagged known disease proteins.
Domain occurrence, repeats and disease statistics
The website features pre-constructed pages that show the database entries sorted according to non-disease and disease-causing proteins respectively. This distinction is applied to the sequence classifications above, the domain occurrence (e.g. listing all domains, Figure 1B), and domain repeats (Figure 1C). This allows database entries to be grouped and examined according to whether the polyQ tracts are found in non-disease or disease-causing proteins (Figure 2).
Figure 2.
Selecting the ‘Stats’ menu text shows the entire database contents to be grouped into non-disease and disease causing proteins. To aid analysis specific entries can be selected (indicated by a tickbox), using the ‘examine’ button and grouped together.
CONCLUSIONS AND FUTURE DIRECTIONS
PolyQ is a valuable resource for theoreticians and experimentalists looking for insights into the context of PolyQ repeats in proteins and relationships with disease. Although the query tool allows searching across much of the database, we are developing a custom interface that will allow user-configurable queries against the whole data set as well as user customization of how the results are displayed. We are also adding the structural information [e.g. from the SCOP (24), CATH (25) and PDB databases (26)] to the resources such that the structural context of polyQ repeats can be investigated.
FUNDING
This work is supported by National Health and Medical Research Council and the Australian Research Council. S.P.B. and A.M.B. are NHMRC Senior Research Fellows. Funding for open access charge: National Health and Medical Research Council (Australia).
Conflict of interest statement. None declared.
REFERENCES
- 1.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S, Berthelier V, Hamilton JB, O'Nuallain B, Wetzel R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry. 2002;41:7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- 3.Robertson AL, Horne J, Ellisdon AM, Thomas B, Scanlon MJ, Bottomley SP. The structural impact of a polyglutamine tract is location-dependent. Biophys. J. 2008;95:5922–5930. doi: 10.1529/biophysj.108.138487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 5.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 6.Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J. Biol. Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 7.Ellerby LM, Andrusiak RL, Wellington CL, Hackam AS, Propp SS, Wood JD, Sharp AH, Margolis RL, Ross CA, Salvesen GS, et al. Cleavage of atrophin-1 at caspase site aspartic acid 109 modulates cytotoxicity. J. Biol. Chem. 1999;274:8730–8736. doi: 10.1074/jbc.274.13.8730. [DOI] [PubMed] [Google Scholar]
- 8.Ellisdon AM, Thomas B, Bottomley SP. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J. Biol. Chem. 2006;281:16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 9.Saunders HM, Bottomley SP. Multi-domain misfolding: understanding the aggregation pathway of polyglutamine proteins. Protein Eng. Des. Sel. 2009;22:447–451. doi: 10.1093/protein/gzp033. [DOI] [PubMed] [Google Scholar]
- 10.Robertson AL, Bottomley SP. Towards the treatment of polyglutamine diseases: the modulatory role of protein context. Curr. Med. Chem. 2010;17:3058–3068. doi: 10.2174/092986710791959800. [DOI] [PubMed] [Google Scholar]
- 11.Faux NG, Bottomley SP, Lesk AM, Irving JA, Morrison JR, de la Banda MG, Whisstock JC. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15:537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto J, Watanabe M, Ichikawa Y, Yee SB, Ihara N, Endo K, Igarashi S, Takiyama Y, Gaspar C, Maciel P, et al. Machado-Joseph disease gene products carrying different carboxyl termini. Neurosci. Res. 1997;28:373–377. doi: 10.1016/s0168-0102(97)00056-4. [DOI] [PubMed] [Google Scholar]
- 13.Padiath QS, Srivastava AK, Roy S, Jain S, Brahmachari SK. Identification of a novel 45 repeat unstable allele associated with a disease phenotype at the MJD1/SCA3 locus. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;133B:124–126. doi: 10.1002/ajmg.b.30088. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Serpell LC, Carter WJ, Rubinsztein DC, Huntington JA. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J. Biol. Chem. 2006;281:15916–15922. doi: 10.1074/jbc.M511007200. [DOI] [PubMed] [Google Scholar]
- 15.Ohshima K, Kang S, Wells RD. CTG triplet repeats from human hereditary diseases are dominant genetic expansion products in Escherichia coli. J. Biol. Chem. 1996;271:1853–1856. doi: 10.1074/jbc.271.4.1853. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar PS, Chang HC, Boudi FB, Reddy S. CTG repeats show bimodal amplification in E. coli. Cell. 1998;95:531–540. doi: 10.1016/s0092-8674(00)81620-7. [DOI] [PubMed] [Google Scholar]
- 17.Margolis RL, Abraham MR, Gatchell SB, Li SH, Kidwai AS, Breschel TS, Stine OC, Callahan C, McInnis MG, Ross CA. cDNAs with long CAG trinucleotide repeats from human brain. Hum. Genet. 1997;100:114–122. doi: 10.1007/s004390050476. [DOI] [PubMed] [Google Scholar]
- 18.Mizutani A, Matsuzaki A, Momoi MY, Fujita E, Tanabe Y, Momoi T. Intracellular distribution of a speech/language disorder associated FOXP2 mutant. Biochem. Biophys. Res. Commun. 2007;353:869–874. doi: 10.1016/j.bbrc.2006.12.130. [DOI] [PubMed] [Google Scholar]
- 19.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Wittkop T, Baumbach J, Lobo FP, Rahmann S. Large scale clustering of protein sequences with FORCE -a layout based heuristic for weighted cluster editing. BMC Bioinformatics. 2007;8:396. doi: 10.1186/1471-2105-8-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C-Y, Chung W-C, Su C-T. Exploiting homogeneity in protein sequence clusters for construction of protein family hierarchies. Pattern Recogn. 2006;39:2356–2369. [Google Scholar]
- 24.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited–architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]