Abstract
The Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis (CAMERA, http://camera.calit2.net/) is a database and associated computational infrastructure that provides a single system for depositing, locating, analyzing, visualizing and sharing data about microbial biology through an advanced web-based analysis portal. CAMERA collects and links metadata relevant to environmental metagenome data sets with annotation in a semantically-aware environment allowing users to write expressive semantic queries against the database. To meet the needs of the research community, users are able to query metadata categories such as habitat, sample type, time, location and other environmental physicochemical parameters. CAMERA is compliant with the standards promulgated by the Genomic Standards Consortium (GSC), and sustains a role within the GSC in extending standards for content and format of the metagenomic data and metadata and its submission to the CAMERA repository. To ensure wide, ready access to data and annotation, CAMERA also provides data submission tools to allow researchers to share and forward data to other metagenomics sites and community data archives such as GenBank. It has multiple interfaces for easy submission of large or complex data sets, and supports pre-registration of samples for sequencing. CAMERA integrates a growing list of tools and viewers for querying, analyzing, annotating and comparing metagenome and genome data.
INTRODUCTION
Metagenomics, termed a new science in a National Academy of Sciences (1) report, is the study of microbial communities sampled directly from their natural environment without prior culturing. Such an approach enables the exploration of the relationships between microbes and their communities and habitats at the most fundamental genomic level. The next generation sequencing technologies like 454 pyrosequencing, Illumina, SOLiD generate large amount of sequencing data in a short time with affordable costs. The data from complexity community sometimes contain more than 10 000 species, but the sequencing reads are sometimes partial fragments of the genomes and low quality reads (2). There are many computional challenges to remove artifacts, annotate and analyze (3) them. The aim of CAMERA is to develop and maintain a rich, distinctive data repository with associated bioinformatics tools, and a collaborative on-line environment; thus, CAMERA is a cyberinfrastructure resource that allows the community to address many of the unique challenges in metagenomics (4) as well as the capacity to share and forward sequence data, metadata and annotations to other resources. CAMERA integrates metagenomic and reference genome projects in four ways: (i) Transferring data directly from a sequencing center. CAMERA has collaborated with the J. Craig Venter Institute (JCVI) for the Global Ocean Sampling (GOS) project and community selected 177 marine microbial reference genomes, Penn State University (PSU) for 226 marine metagenome samples; the Broad Institute for 248 phage genomes, phage metagenomes (viromes) and other ongoing metagenomic projects. (ii) Community or user contributions. (iii) Data exchange with public data resources like NCBI (5), IMG/M (6) and MG-RAST (7). (iv) Integration of reference data sets NCBI, COG (8) and KEGG (9).
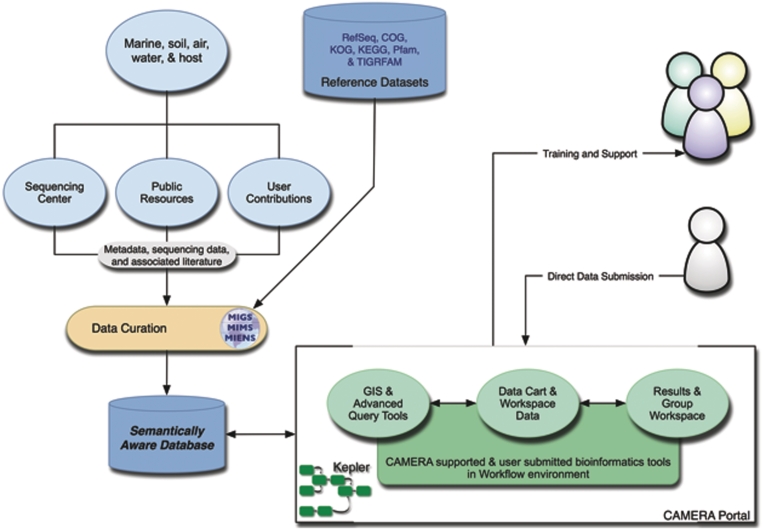
CAMERA launched its website with the GOS data sets and BLAST capability in 2007; then added 155 community selected microbial genomes. The current version of CAMERA, 2.0, was fully released in June 2010 with the following list of key features: (i) Collects and links metadata relevant to environmental metagenome data sets. (ii) Provides facilitation for community data and GSC metadata submission. (iii) Presents the tools for querying metadata across multiple geospatial locations. (iv) Supports advanced queries on environmental parameters and expanded metadata. (v) Exports search results as usable data sets. (vi) Offers large computational and data resources to support analyses with storage of results. (vii) Makes annotation pipelines (workflows) accessible with potential customization and maintains provenance database. (viii) Allows the integration of a diversity of software tools to discover new relationships in microbial ecology research. (ix) Maintains a collaborative group environment, with group access to shared data and analysis results. See Figure 1 for more information. The data and tools are accessible at http://camera.calit2.net.
Figure 1.
Database and data flow.
DATABASE DESIGN AND STRUCTURE
The Semantically-aware Database (SDB) is an integrated database implemented in Oracle to support CAMERA. It is a highly scalable database that contains a broad range of data organized in different modules. These modules include sample metadata, metagenomic sequences (reads, ORFs, proteins, assemblies) and annotations, reference genomes and proteins, and reference databases such as NCBI Taxonomy, Pfam (10) and Go (11). In the database design, we adopted and extended the Minimum Information about a Metagenomic Sequence (MIMS) and Minimum Information about a Genomic Sequence (MIGS) standard established by the Genomics Standards Consortium (12) to manage sample metadata. The metadata entered into the SDB are curated and standardized to support advanced metadata queries.
As the amount of metagenomic and genomic data increases; the SDB is designed to track these changes in the sequence domain. SDB’s reference sequences are refreshed every 2 months following the release cycles of major sequence data sources like NCBI. SDB also has a versioning system implemented so that users can track back to a given prior time what sequences were in the database.
DATA CONTENT
CAMERA contains microbial metagenome data, selected reference marine microbial reference genomes, phage genomes and diverse data from reference resources such as COG, Pfam, KEGG and Genbank Refseq. As of August 2010, it has integrated 72 metagenomic and genomic projects, which include one collection of community selected marine microbial reference genomes and 71 metagenomic projects involving over 800 data sets (>48 billion base pairs, 120 million reads) (visit http://camera.calit2.net for project details). Twenty-one projects involving 192 samples were generated using shotgun sequencing technology, 51 projects involving 399 samples were from 454 pyrosequencing technology. Two projects had both shotgun libraries and 454 libraries. Depending on the level of data analysis, CAMERA hosts raw sequencing reads, assembled contiguous fragments (contigs) and predicted ORFs using six reading frame translation from both reads and contigs. All data sets are searchable as individual project and aggregated data sets.
CAMERA metagenome project data come from various habitats: marine (33 projects), soil (9 projects), fresh water (3 projects), waste water (3 projects), hot spring (3 projects), animal host (11 projects) including the Human Microbiome Project (HMP) and 9 other habitat projects. Over 70 publications are currently linked to samples and projects. Many of the data sets are still being analyzed using CAMERA tools, and/or are still under an embargo period. A large collection of data currently under embargo will be released to public at end of 2010 leading to a doubling of the CAMERA archive during the first quarter of 2011.
DATA QUERY TOOLS
The data is accessible to browse, select, search, download and share using the Geographic Information System (GIS) Query for geographical analysis or using the Advanced Data Query tools for semantic analysis. The GIS Query tool gives users the choice of displaying the map using their preferred base image [Moderate-resolution Imaging Spectroradiometer (MODIS), Aggregate Sea Surface Temperature (SST), CHLO or POC, SODA-POP Model Velocity] and region (EEEs, LME, Longhurst provinces and World seas). It allows the extraction of interpolated values for several physicochemical and biological parameters, such as elevation at sample site or sample depth. Users can view the sample name, location and all other metadata by selecting a sample location on the map; users also can select a custom region by drawing a polygon around the area to be exported. The selection is summarized using a pie chart color coded by projects.
The Advance Data Query interface allows users to browse the database through a hierarchy of projects and or to query the database with specific searches selecting for any of the recorded metadata. The database can be browsed via the following categories: Projects, Samples, Sequence Assays (Libraries), Assemblies or Publications. ‘Projects’ contains the project name, description and data owner’s contact information. ‘Samples’ contains the information on sample collection location, time, pH, habitat, sample depth and other environmental parameters; the sample processing method such as filter size and type, treatment. ‘Sequence Assays’ includes the sequencing reads, sequencing method (Sanger, 454, Illumina or others), template type (cDNA, gDNA or rRNA) and insert size for Sanger reads. ‘Assemblies’ includes the list of assembled libraries and method. ‘Publications’ embraces all the publicly available literature related to projects hosted at CAMERA. The database can also be searched by any metadata category via either a basic search or an advanced search. A basic search is made by selecting a search category. The search categories are the same categories as in the ‘Browse’ section. After selecting a category, users can select a specific search field related to the category, and will then get a ‘Value’ box to enter a search phrase. The default search result is a list of projects. The output can be changed by making a selection in the ‘Output Category’ menu. Advanced Searches are available by adding ‘More Options’. This gives users options for modifying the existing search parameters. For example, clicking ‘More Options’ will allow users to ‘Add’ or ‘Remove’ a search filter. Users can also choose whether to use an ‘AND’ or an ‘OR’ for the new filter. Choosing to ‘Add’ a search filter will give another Search Filter that functions just like the first one. There are a maximum of five total search filters. Clicking on ‘Remove’ will remove the last added filter.
Once users find data that they want to analyze from a GIS or Advance Data query, they are able to store the selected data for further analysis or storage (Figure 2) simply by adding it to a logical data container, which is termed a ‘Data Cart’. A User’s Data Cart stores the information about the data exported from the GIS or Advanced Data query. Within each cart, users can see its contents, which can then be individually or batch exported to the user workspace. The carts are persistent in the system until users delete them. Data that has been exported to a user’s workspace can be used as input for further Data Analysis, and can also be downloaded to user’s local computer.
Figure 2.
Collaborative environment.
DATA SUBMISSION
CAMERA supports the microbial ecology research community by providing a rich, distinctive data repository. To enhance this repository, the CAMERA environment provides users with the ability to share their data with the broader scientific community: (i) the data submission portal was designed to meet the expectations from both experimental and bioinformatics researchers; (ii) a simple submission process (web based entry or template upload); (iii) support from CAMERA staff during the process; (iv) a choice of interfaces to permit a large variety (metadata) and/or quantity of data to be entered readily as well; (v) support for the pre-registration of samples by sequencing centers; (vi) capacity to forward data to other metagenomic sites and data archives such as GenBank; and (vii) submissions must comply with the MIMS/MIGS core, but any metadata can be entered via keywords and free text.) There are different metadata submission forms for different habitats (such as water, soil, air and host)
The ‘Data Submission’ tool is used when researchers would like to contribute their metagenomic sequences and associated metadata to CAMERA for use by the greater scientific community. The data submission page is divided into two main panels: on the left is an overview panel and on the right is a panel that guides the researcher through the submission process. The overview panel contains one tab that shows all the submissions that the researcher has in progress and also, the current stage of these submissions in the submission pipeline. The submission process is composed of three well defined stages. The first stage gathers general project information in order to create the submission; the second stage gathers the sample metadata associated with the sequences; the third stage allows for the upload of the sequence data. CAMERA provides an NCBI upload capability, such that submitted data sets can be forwarded to NCBI for inclusion in GenBank.
DATA ANALYSIS
CAMERA is committed to encouraging and enabling data sharing within the metagenomics community, and provides unique data selection tools allowing users to search and select data both easily and flexibly. However, as the field of metagenomics expands, sharing not just the underlying data itself but also the new and improved analysis methods being developed in the community are ever more important. To that end, CAMERA 2.0 supports an advanced data sharing environment along with a collaborative analysis environment that provides an extensible collection of bioinformatics tools and workflows to address the unique challenges of metagenomics and enables researchers to collaborate in new ways through CAMERA.
CAMERA 2.0 organizes data analysis tools through a collaborative scientific workflow system. At the core of this environment is the Kepler scientific workflow system (13, 14) that supports integration of data and automated computational tools while also providing capabilities for the tracking of provenance, i.e. recording all steps and features associated with the processing of data. This data-oriented view of an analysis, captured via end-to-end provenance of a workflow, enables the communication of what has occurred within a workflow to collaborators, and also, allows for the exchange and reproducibility of the computation itself. Through the CAMERA portal, users can create, share, retrieve and run the processing workflows specific to their own experiment without having to install special software. In addition, this workflow-based environment reduces the cost of moving from a stand-alone scientific application to a workflow-based community resource by (i) designing and publishing workflows based on application services; (ii) executing workflows based on local or online data; (iii) saving and querying workflow results; (iv) saving and viewing data and its provenance; (v) creating ad-hoc collaborations and project spaces; and (vi) publishing uploaded workflows or sharing workflows with collaborative group members.
Currently, the core workflow system makes the following metagenomic analyses available to researchers: data quality control (specifically, QC Filter and 454 Duplicate Clustering), read assembly (454 Read Assembly), functional annotation and clustering (Metagenomic Data Annotation and Clustering), BLAST and additional downstream analysis methods. The QC Filter takes fasta and qual files or a fastq file as input, calculates the average score for each read, then fetches high quality reads, filters out reads shorter than the minimum read length and generates a statistical analysis. The 454 Duplicate Clustering identifies exact and near identical duplicates to remove sequencing artifacts (15). The 454 Read Assembly first runs seven independent assembly programs to get a pool of contigs, then re-assembles these contigs to create a consensus—this re-assembly process significantly captures the benefit of all individual assemblers. The Metagenomic Data Annotation and Clustering workflow (16,17) identifies tRNAs, rRNAs and ORFs from the input reads, performs clustering on the reads and ORFs, and then annotates against Pfam, TIGRFAM and COG. The BLAST module offers all six BLAST programs, takes as many as several hundred thousand sequences and runs these against metagenomic data sets or reference genomes, and has a graphic output interface as well as export functions.
Another important aspect of the workflow environment is that these workflows are organized into a systematic network, in which the output for one functional unit can be used as an input for the next workflow. This allows researchers to build a complete end-to-end analysis stream by choosing to use different combinations of workflows based on their specific needs for that data analysis. For example, one possible end-to-end analysis stream for researchers with raw sequencing data might entail: (i) using the QC filter to do data quality control; (ii) assembling the resultant reads to longer contigs; (iii) assigning taxonomy to each of these contigs; (iv) annotating genes against COG, Pfam, TIGRFAM and other reference databases and then clustering the genes to a desired level; and (v) running a statistical comparison, obtaining a graphic view, and other analyses. The workflow linkage capability provides researchers with the convenience of ‘one stop shopping’. All the results from workflow runs are accessible to users through graphic views and /or bulk downloads of the processed and raw outputs. Users have a choice to analyze results either by CAMERA cutoffs or by custom values.
NEAR TERM EXTENSIONS
A taxonomy binning workflow, which will assign a taxonomy path to a read using tools like MEGAN (18), RDP Classifier (19) and the Silva database (20) will be released at the end of 2010. To enhance studies on microbial diversity and relationships, phylogenetic and single gene analylsis workflows are in the process of development and will be released in the second quarter of 2011. New visualization tools are currently being developed in order to improve the efficiency of analyzing large and complex metagenome data sets that are generated with new technology platforms, such as 454 Life Science, Solexa and SOLiD. CAMERA is also integrating workflows for specified domain areas from the research community and will establish opportunities for the community to incorporate their own novel software into the workflows.
FUNDING
Gordon and Betty Moore Foundation; National Center for Research Resources (NCRR), National Institutes of Health (grant R01RR025030); National Science Foundation SDCI Award (OCI-0722079 to Kepler/CORE); Department of Energy grant (DE-FC02-01ER25486 to SciDAC/SDM). Funding for open access charge: Gordon and Betty Moore Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Handelsman J, Tiedje JM, Alvarez-Cohen L, Ashburner M, Cann IKO, Delong EF, Doolittle WF, Fraser-Liggett CM, Godzik A, Gordon JI, et al. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet. Washington, DC: The National Academies Press; 2007. [PubMed] [Google Scholar]
- 2.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput. Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wooley JC, Ye Y. Metagenomics: facts and artifacts, and computational challenges. J. Comput. Sci. Technol. 2010;25:71–81. doi: 10.1007/s11390-010-9306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, Chen IMA, Grechkin Y, Dubchak I, Anderson I, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acid Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The Metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatusov RL, Koonin EV, Lipman DJ. A Genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GO-EBI, EMBL-EBI. The gene ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38(Suppl. 1):D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deelman E, Gannon D, Shields M, Taylor I. Workflows and e-science: an overview of workflow system features and capabilities. FGCS. 2009;25:528–540. [Google Scholar]
- 14.Ludaescher B, Altintas I, Berkley C, Higgins D, Jaeger E, Jones M, Lee EA, Tao J, Zhao Y. Scientific workflow management and the Kepler system. Concurrency Comput. Pract. Exp. 2006;18:1039–1065. [Google Scholar]
- 15.Niu B, Fu L, Sun S, Li W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics. 2010;11:187. doi: 10.1186/1471-2105-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics. 2009;10:359. doi: 10.1186/1471-2105-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Gilna P, Li W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics. 2009;25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]