Abstract
dictyBase (http://www.dictybase.org), the model organism database for Dictyostelium, aims to provide the broad biomedical research community with well integrated, high quality data and tools for Dictyostelium discoideum and related species. dictyBase houses the complete genome sequence, ESTs, and the entire body of literature relevant to Dictyostelium. This information is curated to provide accurate gene models and functional annotations, with the goal of fully annotating the genome to provide a ‘reference genome’ in the Amoebozoa clade. We highlight several new features in the present update: (i) new annotations; (ii) improved interface with web 2.0 functionality; (iii) the initial steps towards a genome portal for the Amoebozoa; (iv) ortholog display; and (v) the complete integration of the Dicty Stock Center with dictyBase.
INTRODUCTION
Dictyostelium offers unique opportunities to study gene function and conserved biological processes in a simple model system. As one of the earliest branches to emerge after the plant and animal split, Dictyostelium provides invaluable insights into the basic biology of eukaryotes (1,2). This also makes it uniquely valuable for comparative genomics studies. dictyBase is the manually annotated model organism database for D. discoideum (3). It contains the entire 34 Mb nuclear genome sequence of the commonly used haploid laboratory strain, AX4 (1) the 55-kb mitochondrial genome (4), the extrachromosomal ribosomal RNA genes (5) and over 162 000 EST sequences (6). Since 2010 dictyBase has also housed the D. purpureum genome in a database that uses the dictyBase infrastructure (Sucgang,R. et al., submitted for publication).
The D. discoideum genome is manually annotated at dictyBase. All literature describing genes from this organism is integrated in the database and used to annotate gene product functions, strains and mutant phenotypes, and to associate gene ontology terms with gene products.
In this report we describe new data and tools since our last update in 2009 (7) including new annotations; improved interface with web 2.0 functionality; early steps towards a genome portal for the Amoebozoa; orthology display; and the complete integration of dictyBase with the Dicty Stock Center.
NEW ANNOTATIONS
Gene model curation has been a priority since the inception of dictyBase. Each automated gene prediction is inspected by a curator who reviews supporting data, such as ESTs and sequence similarity to other species. Gene models are corrected as necessary and promoted to a curated model based on available experimental data. Between 15% and 20% of the computational gene predictions require manual correction. To accelerate the rate of gene model curation, we established a prioritization system taking into account: (i) the amount and types of data associated with a gene, such as ESTs, RNA sequencing data and homologous sequences, and (ii) whether there was agreement between two sets of automated gene predictions, the prediction from the sequencing project annotation pipeline (i), and (ii) an in-house gene prediction we have run based on our fully supported curated gene models. Genes with high level of support and gene predictions confirmed by the two methods were reviewed first. A high fraction of those gene models were correct and approximately 1000, or 30% of the genes that were left to annotate, were manually approved in a very short time span (about a month). We have also developed a new gene curation tool that presents the curator with all available information to make a gene model: sequence and gene coordinates (including exon/intron boundaries); expression information (ESTs and RNA seq), alignment of protein sequences with those of its closest sequenced genetic neighbors, D. purpureum, D. fasciculatum and P. pallidum, and two automatically predicted gene models.
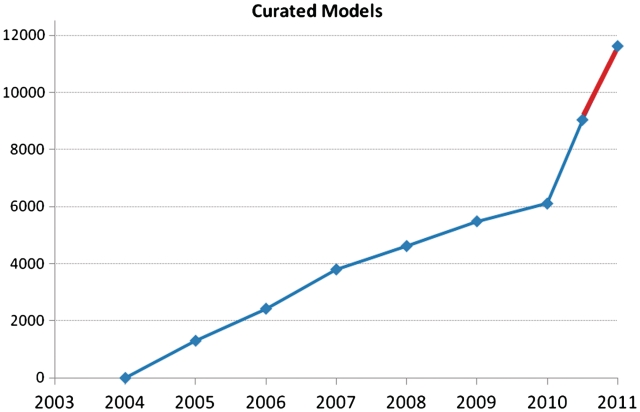
We are now in the process of reviewing genes for which there is less support and have annotated 2845 genes since we have started working from those priority lists. According to our most recent estimates, the Dictyostelium genome contains 12 646 protein coding genes, therefore, we have less than 2000 genes models still to be manually reviewed, assuming that there are 1000 genes lacking any supporting data and that we will not be able to verify. Using this new technology and prioritization, we estimate that a first pass of all gene model annotation will be completed by early 2011 (Figure 1).
Figure 1.
Gene model curation progress since 2003. The new curation strategy has resulted in a 10-fold increase in the rate of gene model curation from April to August 2010 compared with the previous years. The completion of a first pass of gene model curation, shown in red, is predicted for the end of 2010.
Another important activity of dictyBase curators is to annotate genes with the data from the nearly 7000 references mentioning Dictyostelium present in PubMed, 1750 of which have been curated. Those annotations include function descriptions, gene ontology terms, as well as strains and phenotypes. We have also annotated nearly 500 transposable elements (named with RTE and TE suffixes) with the valuable assistance of Thomas Winckler (University of Jena). An overview of the annotation coverage of the Dictyostelium genes is shown in Table 1.
Table 1.
Data and annotations in dictyBase as of September 2010. Total numbers for each category is shown in bold on the last line
| Curated model | Gene Ontology | Phenotype | Number of genes |
|---|---|---|---|
| + | + | + | 716 |
| + | + | − | 6,964 |
| + | − | + | 764 |
| + | − | − | 1001 |
| − | − | − | 3201 |
| 9445 | 7608 | 764 | 12646 |
IMPROVED DISPLAY AND WEB 2.0 FUNCTIONALITY
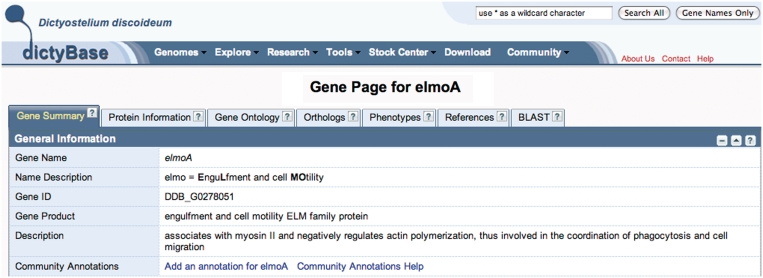
We have modernized the dictyBase interface to use Web 2.0 technology and the YAHOO User Interface library to enhance the user interface of dictyBase. We have reengineered our gene page to display different types of information, such as gene ontology, phenotypes, references and protein information in separate tabs on the gene page (Figure 2). The gene summary tab contains general information, including gene name, synonyms, gene product names and a short description of the gene product’s function. It also includes sections with genomic coordinates and sequence information as well as an overview of all annotations. In cases where a gene encodes more than one transcript, the gene product section displays sub-tabs for each splice variant. The reengineered page also displays protein information obtained from UniProtKB as well as InterPro protein domains, displayed both graphically and in tabular form. A phenotype tab lists all strains relevant to the gene with their mutant characteristics and phenotype information. Strain availability in the dictyBase stock center is indicated by a clickable green basket that can be used to initiate ordering of strains. The gene ontology annotations, complete references and a BLAST server are also accessible from individual tabs.
Figure 2.
The updated dictyBase gene page. Information is organized in tabs. The annotations are also present on the main gene page in an abbreviated form.
In addition to the new organization of the gene page, the Web 2.0 framework is built to allow parallel processing for rendering of different sections of the page. This parallel rendering combined with a caching scheme drastically improved the speed of loading a gene page, even though a large amount of data is being displayed.
TOWARDS A GENOME PORTAL FOR AMOEBOZOAN SPECIES
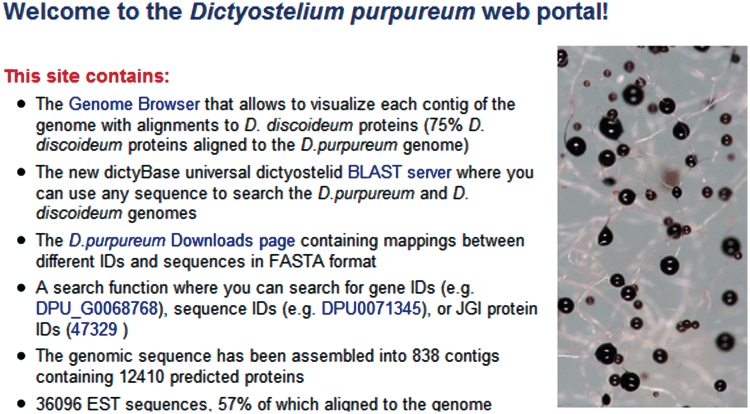
The genome sequences of several Dictyosteliid species are now available or will soon become available. This data is extremely valuable to help better define the gene models and in evaluating conserved elements across this evolutionarily diverse clade of organisms. In February 2010, we released the D. purpureum genome at dictyBase (http://genomes.dictybase.org/purpureum; Figure 3). The D. purpureum site has the same ‘look and feel’ as the D. discoideum site, where each gene has its own gene page and the contigs are represented graphically in the Generic Genome Browser (8) (contigs are represented rather than chromosomes because the genome assembly is not yet complete). The D. purpureum Genome Browser shows alignments of D. discoideum proteins generated by TBLASTN, hyperlinked to the respective gene page on the D. discoideum site. We are working towards providing similar genome sites for other sequenced amoebozoan species, including D. fasciculatum and Polysphondylium pallidum.
Figure 3.
The D. purpureum genome portal. The front page of the D. purpureum genome portal contains a description and links to the contents of the site.
We have updated the dictyBase BLAST server to provide access to the gene/protein sequences as well as ESTs of D. purpureum (submitted for publication), and gene/protein sequences of D. fasciculatum, and P. pallidum (generously provided by Gernot Glöckner). A ‘BLAST-All’ option allows simultaneous queries of sequences from all available organisms. The dictyBase BLAST server will continue to expand as sequences from different species become available.
ORTHOLOG DISPLAY
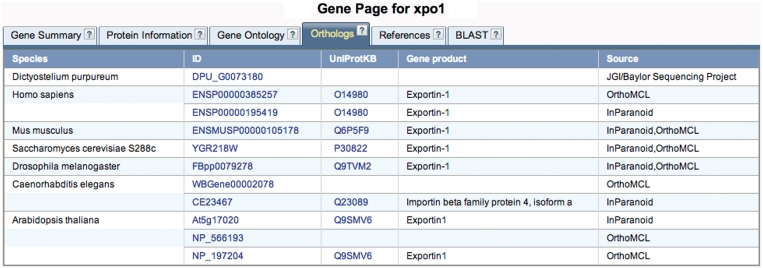
An important application of research using model organisms is to provide insight about conserved biological processes. To make maximal use of the knowledge gained using Dictyostelium, it is very important to be able to compare the known functions of genes with their counterparts from other species and vice versa. To help facilitate those analyses, dictyBase gene pages include a new tab with orthologs of eight different species: D. purpureum (and conversely D. discoideum orthologs on the D. purpureum gene page), Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, Arabidopsis thaliana and Escherichia coli (an example is shown in Figure 4). Ortholog data was obtained from InParanoid [inparanoid.sbc.su.se, (9)] and OrthoMCL [orthomcl.org, (10)], and in the case of D. purpureum from A. Kuspa (private communication). The data is shown in a table containing the species name, a link to the sequence used to calculate the orthologs (usually the model organism database for the species, or ensembl), a link to UniProt (when available), and the gene product name. It should be noted that InParanoid and OrthoMCL calculate both orthologs and paralogs.
Figure 4.
Ortholog display on the gene page. Ortholog data is shown in a new ‘Orthologs tab’ from different sources: OrthoMCL, InParanoid and D. purpureum orthologs from A. Kuspa (personal communication). Links to the original data used to generate the ortholog groups are shown in the column labeled ‘ID’; as well as links to UniProtKB.
The complete lists of orthologs are also available as a text file on the dictyBase Downloads site.
INTEGRATION OF THE DICTY STOCK CENTER AND dictyBase
The Dicty Stock Center currently holds over 1500 strains targeting over 930 different genes. There are over 100 different distinct amoebozoan species. In addition, the collection contains nearly 600 plasmids and other materials such as antibodies and cDNA libraries. The Dicty Stock Center receives about one order a day ranging from one to about 50 items. We have shipped over 1500 individual items to 17 different countries since March 2009. The strain and plasmid collection continue to expand, as we request new strains upon publication. We send a weekly newsletter with the list of materials newly received to dictyBase users.
dictyBase has been supporting the bioinformatics infrastructure of the Dicty Stock Center since its inception, and, as of March 2009, the stock center is operated from Northwestern University together with dictyBase. This tighter integration of the two resources has improved curation consistency and streamlined the strain collection process. The Dicty Stock Center is highly valued by the research community, and together with the genome database, has been instrumental in attracting new groups to Dictyostelium as a model system for biomedical research.
CONCLUSION/FUTURE DIRECTIONS
Genomic and proteomic technologies are rapidly improving, resulting in rapid increases in the amount of high quality large-scale data that are produced by researchers. The ability of investigators to use the data meaningfully is highly dependent on robust interfaces, efficient search technology and data management that is best housed at a community resource such as dictyBase. We will continue to incorporate new data as it becomes available, including sequence and gene expression data from large-scale genomics projects. This includes: genome sequences of several species related to D. discoideum and nucleotide polymorphism data from other D. discoideum strains, in particular NC4, the wild-type parent of all laboratory strains. Several groups are planning to share RNA sequence data from wild-type and mutant cells, and in different physiological conditions (cell cycle, development). Finally, we have established a collaboration with IntAct at the EBI (11) to capture protein-protein interaction data.
FUNDING
National Institutes of Health GM64426, GM087371 and HG0022 (to dictyBase and the Dicty Stock Center). Funding for open access charge: Northwestern University.
Conflict of interest statement. None declared.
Box 1. Data availability.
The data and annotations from dictyBase are accessible though a general search tool that searches gene names, gene product names, gene descriptions, Gene Ontology terms, dictyBase gene ID (DDB_G###), dictyBase sequence IDs (DDB###), ESTs, gene descriptions, plasmids, strains, phenotypes, GenBank accession numbers, authors, colleagues and web pages. We also have an implementation of BioMart (13), accessible at http://dictybase.org/biomart/martview.
dictyBase provides all of its data as bulk downloads available from http://dictybase.org/Downloads/. This data is available in multiple formats including excel spreadsheets, tab-delimited formats. Data currently available includes all dictyBase gene sequences and annotations in GFF3 format, sequence information in FASTA format, curated model history as tab-delimited files, all curated strain information (excel and tab-delimited), all gene IDs, names, synonyms and gene product terms. Each of these files is updated regularly to assure they contain the most current information. In addition to these bulk data sets, dictyBase regularly deposits updated sequence information and annotations with GenBank to assure this information is widely available from NCBI, as well as from sites such as UniProt/KB that use GenBank as a data source. We are also directly sharing data with UniProt/KB and several other resources such as Gene Ontology Consortium, Ensembl Genomes (Ensembl protists), orthology resources such as InParanoid, as well as many other informatics resources.
To provide a way for users to map different IDs, we have developed an ID converter tool. The tool inter-converts sequence IDs (DDB# or UniProt IDs) and gene IDs (DDB_G#) IDs. It provides outputs in plain text and Excel formats. This allows researchers to efficiently link their studies to the most current dictyBase identifiers.
In addition to data, all of our software and tools are also available. We have developed Modware for GMOD as an object oriented API for the Chado database schema. It is available at http://gmod-ware.sourceforge.net/.
REFERENCES
- 1.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, Cavender J, Milano-Curto A, Rozen DE, Dingermann T, et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chisholm RL, Gaudet P, Just EM, Pilcher KE, Fey P, Merchant SN, Kibbe WA. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34:D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa S, Yoshino R, Angata K, Iwamoto M, Pi M, Kuroe K, Matsuo K, Morio T, Urushihara H, Yanagisawa K, et al. The mitochondrial DNA of Dictyostelium discoideum: complete sequence, gene content and genome organization. Mol Gen Genet. 2000;514:519–541. doi: 10.1007/pl00008685. [DOI] [PubMed] [Google Scholar]
- 5.Sucgang R, Chen G, Liu W, Lindsay R, Lu J, Muzny D, Shaulsky G, Loomis W, Gibbs R, Kuspa A. Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res. 2003;31:2361–2368. doi: 10.1093/nar/gkg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urushihara H, Morio T, Tanaka Y. The cDNA sequencing project. Methods Mol. Biol. 2006;346:31–49. doi: 10.1385/1-59745-144-4:31. [DOI] [PubMed] [Google Scholar]
- 7.Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, Merchant SN, Bushmanova YA, Shaulsky G, Kibbe WA, et al. dictyBase–a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–D519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berglund AC, Sjolund E, Ostlund G, Sonnhammer EL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]