Abstract
Coronary artery disease (CAD) is a complex, multifactorial disease and a leading cause of mortality world wide. Over the past decades, great efforts have been made to elucidate the underlying genetic basis of CAD and massive data have been accumulated. To integrate these data together and to provide a useful resource for researchers, we developed the CADgene, a comprehensive database for CAD genes. We manually extracted CAD-related evidence for more than 300 candidate genes for CAD from over 1300 publications of genetic studies. We classified these candidate genes into 12 functional categories based on their roles in CAD. For each gene, we extracted detailed information from related studies (e.g. the size of case–control, population, SNP, odds ratio, P-value, etc.) and made useful annotations, which include general gene information, Gene Ontology annotations, KEGG pathways, protein–protein interactions and others. Besides the statistical number of studies for each gene, CADgene also provides tools to search and show the most frequently studied candidate genes. In addition, CADgene provides cumulative data from 11 publications of CAD-related genome-wide association studies. CADgene has a user-friendly web interface with multiple browse and search functions. It is freely available at http://www.bioguo.org/CADgene/.
INTRODUCTION
Coronary artery disease (CAD), also known as coronary heart disease (CHD), is the most common cause of death in the world. Many research efforts have been made to identify its acquired and inherited risk factors (1). CAD is a complex disease influenced by multiple combinations of gene–gene and gene–environment interactions (2). The heritability of CAD is estimated to be 40 to 60%, however the underlying genetic mechanism is poorly understood (3). CAD occurs when coronary arteries are narrowed or occluded due to stenosis, which reduces or blocks the blood supply through the diseased arteries to the heart. The main cause of narrowing of the coronary artery is the atherosclerotic plaque, while the main cause of the artery occluding is the thrombus usually formed by atherosclerotic plaque rupture (4). There are many hypotheses proposed to explain the underlying mechanisms of the initiation, progression and rupture of the coronary atherosclerotic plaque (5). The key points of these hypotheses include: (i) lipoprotein retention; (ii) endothelial dysfunction; (iii) immune and inflammation response of the artery; (iv) vascular smooth muscle cell (VSMC) proliferation; (v) lipid absorption by macrophage and VSMCs, and the formation of foam cells; (vi) platelet activation and thrombosis (4,6). It is challenging to explore the molecular mechanisms underlying the development of CAD. A systematic CAD gene database will be a valuable and essential resource for the research community.
Over the past decades, many experimental strategies (genome-wide linkage scan, association studies, global microarray gene expression analysis, proteomics, etc.) and large efforts have been applied onto the studies of CAD. A number of genomic regions, variants in candidate genes and risk factors were implicated in increasing the susceptibility of CAD (7). However, most of the variants and genes have not been established consistently. The most robust genetic risk variant for CAD was identified on chromosome 9p21.3 by genome-wide association studies (GWAS) (8). As case–control association studies are widely used to identify susceptibility genes, a large number of candidate gene association studies for CAD were published and massive data has been accumulated. Although the results of some studies conflict with others, these findings did provide important clues to explore the mechanisms in the pathogenesis of CAD. Genes involved in different pathways and functions such as altered lipoprotein handling, disruption in endothelial integrity, arterial inflammation and thrombosis were found to be related to CAD (9). In recent years, as the cost of the high-throughput genotyping decreased rapidly, GWAS are becoming a popular and powerful approach to identify candidate variants for a specific phenotype or disease. Several GWAS for CAD and myocardial infarction (MI) have been conducted, and identified many CAD-associated variants (10,11). Thus, these data provide an unprecedented opportunity to construct a useful gene resource for CAD, which can integrate and analyze these data to explore the pathogenesis of CAD. To date, a few databases for cardiovascular diseases have been developed, such as the Cardio database (12), inherited arrhythmias database (http://www.fsm.it/cardmoc/), CaGE (13) and HuGE Navigator Phenopedia (14). However, none of them are specific to CAD, the most common cardiovascular disease. Some of these databases were not updated for >5 years or even not available at the present time.
Here we present a database for CAD genes (CADgene, http://www.bioguo.org/CADgene/). Aiming to efficiently integrate and analyze all or most of the published CAD-related gene studies, we collected CAD candidate genes and their detailed evidence associated with CAD from publications (SNPs, P-value, odds ratio, etc.). For each gene, we made useful annotations, which include basic gene information (ID, name, alias, location etc.), Gene Ontology (GO) annotation (15), KEGG pathway (16) and protein–protein interaction (PPI) information. In this work, we took advantage of the current CADgene data and made some interesting statistical analyses and discussions. CADgene seeks to be a useful resource for the research communities of CAD and other cardiovascular diseases.
DATA SOURCES AND METHODS
Data collection and curation
Currently, CADgene database contains >300 CAD-related genes and their detailed information associated with CAD from >1300 CAD-related publications published before 20 June 2010. To obtain a complete list of publications for CAD genes, we made a comprehensive search for CAD-related studies in NCBI PubMed. We used the following search terms for PubMed searching.
(‘coronary artery disease’ [Title/Abstract] OR ‘coronary heart disease’ [Title/Abstract]) AND (‘gene’ [Title/Abstract] AND (‘association’ [Title/Abstract] OR ‘microarray’ [Title/Abstract] OR ‘expression’ [Title/Abstract] OR ‘linkage’ [Title/Abstract] OR ‘proteomics’ [Title/Abstract] OR ‘metabolism’ [Title/Abstract] OR ‘metabolomics’ [Title/Abstract] OR ‘metabonomics’ [Title/Abstract]))
As a result, we obtained >2000 CAD-related publications, most of which are candidate gene association studies or replication studies of previous findings. After manually scanning the abstracts of these articles, we excluded the reviews and those that studied other diseases instead of CAD. Finally, we retained ∼1300 publications which studied CAD candidate variants or genes. We extracted the CAD-related detailed information for the reported CAD genes from the abstracts by manually reading and curation. The extracted information from a CAD-related article includes: population, case and control, study subjects (gene, SNP, mutation, expression level, etc.), phenotype (CAD or MI), main results and conclusion of the publication. Since we aim to construct a CAD gene database, we organized these data as gene-centered by manually converting different gene names in publications to the unique NCBI Entrez Gene ID (http://www.ncbi.nlm.nih.gov/gene).
As GWAS is a powerful and unbiased strategy for identifying phenotype associated variants, it is necessary to include the available GWAS data in our database. We made a comprehensive search for GWAS of CAD and MI, and obtained 11 publications. We carefully read the full text of these GWAS publications and extracted the significant results. Since the associated SNPs may reside in genic or intergenic regions, we organized the curated GWAS data as SNP-centered instead of gene-centered as used in candidate gene studies.
Functional categories
Based on the proposed mechanisms and pathways for CAD as mentioned above, we classified genes from candidate gene studies in CADgene into 12 categories according to their functions (Table 1). Except the ‘Others’ category, all other 11 functional categories are relatively independent functional pathways related to the development of atherosclerotic plaque or risk factors of CAD. For example, the endothelial dysfunction has been considered as the first step toward atherosclerosis and a common link of all cardiovascular risk factors (17); immune cells dominate early atherosclerotic lesions and atherosclerosis is considered as an inflammatory disease (18). The ‘Others’ category includes genes in other functions rather than in the 11 categories. In our opinion, such classification will help users to browse and understand these data in the CADgene database.
Table 1.
Summary of functional categories in CADgene
| Functional category | Number of genes | Number of articles |
|---|---|---|
| Lipid and lipoprotein metabolism | 53 | 398 |
| Immune and inflammation | 68 | 173 |
| RAS | 11 | 152 |
| Endothelial integrity | 31 | 134 |
| Thrombosis | 26 | 120 |
| Oxidation–reduction state | 22 | 97 |
| Homocysteine metabolism | 7 | 71 |
| Metalloproteinase and ECM | 11 | 24 |
| Gender difference | 6 | 24 |
| Glucose metabolism | 8 | 17 |
| VSMC abnormalities | 7 | 7 |
| Others | 68 | 84 |
Gene and SNP annotation
Besides the collected evidence from the abstracts, we provided as many annotations as possible for each gene to facilitate the interpretation of the relation with CAD, especially the annotations of GO, KEGG pathways and PPIs. We used NCBI Entrez Gene ID or gene symbol as the central ID for cross linking and annotation. We downloaded the basic gene annotation files from NCBI FTP site. We parsed the gene_info and gene2refseq files to retrieve the basic gene information such as gene symbol, name, chromosome, genetic location, gene type and reference sequence information. We obtained the GO annotations for each gene from the gene2go file and downloaded the gene pathway data from KEGG database. We integrated the PPI information from both HPRD (19) and BioGRID (20), the two largest PPI databases, and then extracted the direct interactors for CAD candidate proteins in CADgene. For SNPs, we also provide their basic information including chromosomal location, allele, host genes, etc.
DATABASE CONSTRUCTION AND WEB INTERFACE
Database organization
We stored and managed all the data in MySQL, which is a popular and open source database management system widely used in bioinformatics and biomedical database development. Since candidate gene association studies and GWAS have different data formats and contents, we managed these data by separate tables. The article information, CAD gene evidence extracted from publications, basic gene information, reference sequences, GO annotations, KEGG pathways and PPIs were stored into individual tables.
Data browse
A user-friendly web interface was designed and implemented for CADgene. It is freely available at http://www.bioguo.org/CADgene/. Users can browse or search all the data at different levels.
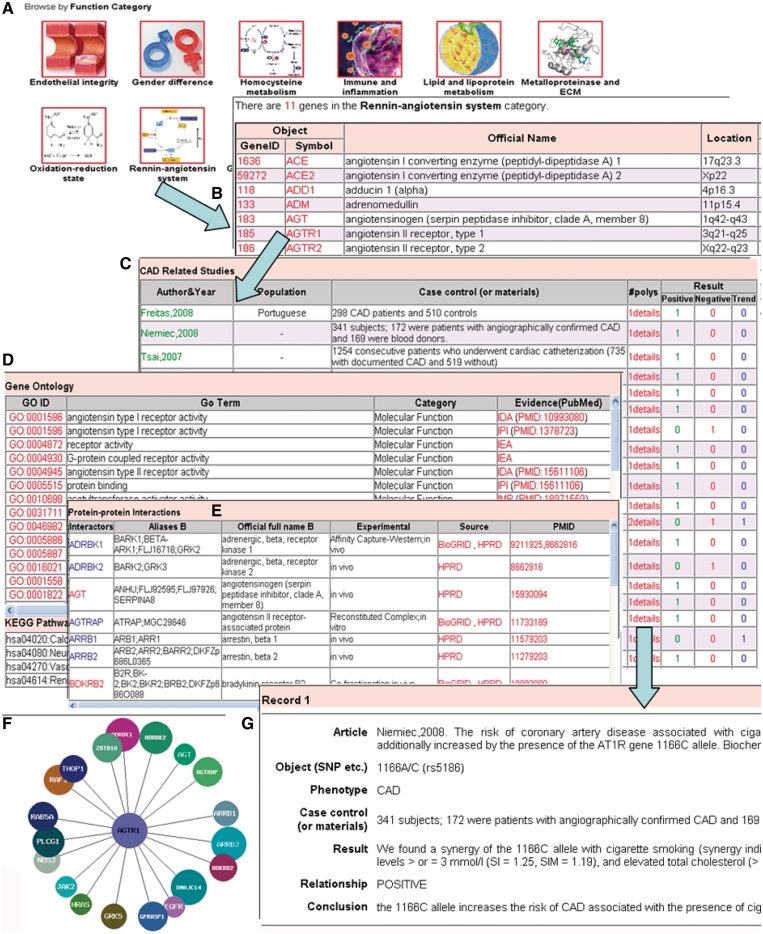
To help users to browse the data conveniently, CADgene provides three different methods for browsing CAD-related genes from candidate gene studies: (i) by functional category; (ii) by chromosome; or (iii) through a summary page which lists all the genes and the summary of their CAD-related studies. A cascading style is applied for data browsing, e.g. from functional category to gene list, and then to gene information (Figure 1). Selecting a functional category will list the CAD candidate genes in this category (Figure 1A and B). When users click a gene name on the gene list page, it will show the gene information page, which contains the summarized information of its CAD-related studies (e.g. author and year, population, case and control, studied polymorphism number and result assessment) and gene annotations (e.g. gene symbol and name, GO annotations, RefSeq, database cross links, KEGG pathways and PPIs) (Figure 1C–F). CADgene provides the PPI information for each protein by both a table list and an interaction network figure. By clicking the hyperlink in the polymorphism column, it will open the detailed page for the study that shows the relation between the gene (SNP) and CAD (Figure 1G). For GWAS data, we made a separate page to browse them, which is similar with candidate gene browsing.
Figure 1.
Examples of the cascading browse and gene information in CADgene. (A) Functional categories in CADgene. (B) Gene list for the RAS functional category. (C) CAD-related studies for a specific gene (i.e. AGTR1). (D) GO and KEGG annotations. (E) PPI information in a table. (F) Graphic show of the PPI network. (G) Detailed information of a CAD-related study.
For further use of the data, we provided an advanced browse tool for CADgene. On the advanced browse page, users can browse data by combination of gene name with the publication date or browse only the statistically significant or non-significant studies for genes. Users may be interested in those genes with multiple significant studies. Advanced browse page provides such a tool to view genes by the number of their studies. To analyze the most frequently studied genes, we designed a page to show the statistical results of those genes with more than10 significant or nonsignificant studies. Additionally, users can browse genes or SNPs identified in both candidate gene studies and GWAS, which may serve as important links between the two kinds of studies.
Data search
CADgene provides three approaches for searching the data, including the text search and sequence search. First, users may find a quick search box on the top right of each page for searching by gene ID or symbol or SNP id. Second, CADgene provides an advanced search page, on which users may search data through the gene name, SNP ID, chromosomal locus, article information or gene annotation. Additionally, we provide a batch search function for a gene list on this page. Third, a BLAST search against the nucleotide or protein sequences of the CAD-related genes is also available in CADgene.
DISCUSSION
The study history of CAD genes
As CADgene contains most of the CAD-related association studies, we roughly summarized the research history of CAD genes by the number of publications and published date (Supplementary Table S1). The earliest study collected in CADgene was published in 1988. Genes encoding apolipoproteins were first shown to be associated with CAD and the lipoprotein metabolism is still the research hotspot for CAD. All the nine studies published before 1990 in CADgene were studies of apolipoproteins (APOA1, APOC3 and APOB). Then, genes in the rennin–angiotensin system (RAS) and Thrombosis categories began to be studied and thereafter genes in all the 12 functional categories were studied. After 2000, the CAD studies developed rapidly and ∼80% studies in CADgene were published in the recent 10 years. Lipid and Lipoprotein Metabolism, Immune and Inflammation and RAS are the top three studied functional categories for CAD. Studies for genes in the top six functional categories account for >80% of all the studies in CADgene (Supplementary Table S1). In the recent 5 years (2006–2010), the studies of Immune and Inflammation, and Endothelial Integrity categories were developed quickly. CAD has been considered as a disease of immune and inflammation (21), and endothelium dysfunction has been considered as the first step toward CAD (17).
Most popular studied genes
To show the most popularly studied CAD genes, CADgene provides a web page to show them (http://www.bioguo.org/CADgene/topGenes.php). The hottest gene is ACE (angiotensin I converting enzyme), a gene in the RAS category, which has more than100 studies in total. There are 25 genes with more than 10 significant studies and five of them also have more than 10 non-significant studies. Because CAD is influenced by multiple genetic and environmental factors, studies with different cases and populations may obtain different results for various pathogenesis. These 25 genes are distributed in eight functional categories and 11 of them are genes in the Lipid and Lipoprotein Metabolism category, which includes five apolipoproteins (APOE, APOB, APOA5, APOC3 and APOA1).
KEGG pathway and PPI
CAD is a complex disease and there may be huge biological networks and pathways contributing to the pathogenesis of CAD. So we provided the KEGG pathway and PPI information for genes in CADgene.
For each candidate gene, CADgene displays its involved KEGG pathways and all the CAD-related genes in these pathways. As a result, 155 and 115 KEGG pathways contain at least one and two genes in CADgene, respectively, which reflects the complexity of CAD. Pathways playing important roles in the pathogenesis of CAD are at the top enriched pathway list, such as cytokine–cytokine receptor interaction (22), complement and coagulation cascades (23), PPAR signaling pathway (24), MAPK signaling pathway (25) and focal adhesion (22) containing 29, 25, 17, 16 and 12 CAD-related genes, respectively (Supplementary Table S2). We suggest that more attention should be paid to the pathways containing multiple CAD-related genes, and also genes involving many pathways because they may act as linkers of these pathways.
In addition to the pathway information of CAD-related genes, CADgene also provides the direct interactors of CAD proteins and their graphic PPI networks. For each protein, we summarized the numbers of its interactors in CADgene and in the human interactome, and the ratio of them. We are interested in the top 21 proteins which have no less than three interactors in CADgene and >50% of their interactors are in CADgene gene list (Supplementary Table S3). For example, ACE is the most studied gene for CAD and all its interactors (AGTR2, BDKRB2 and COMT) are in CADgene with CAD-related studies. We also examined the enriched pathways for interactors of each protein in CADgene. We are interested in the nine proteins accompanied by five or more CAD-related interactors in a specific pathway and listed them in Supplementary Table S4. Five of the nine enriched pathways are the complement and coagulation cascades pathway, which may suggest its importance in CAD. The protein encoded by the gene F2 (also known as coagulation factor II, prothrombin) has 45 interactors totally and 17 of them are CAD-related proteins in CADgene. Among the 17 interactors, 14 are involved in the complement and coagulation cascades pathway. Thus, the gene F2 should be very important in the complement and coagulation cascades pathway and in the CAD-related network. Although the gain-of-function F2 G20210A mutation was proved to be involved in the development of venous thrombosis (26), which may suggest its importance in the complement and coagulation cascades pathway. However, gene F2 was just reported to be associated with CAD at a modest risk (OR = 1.3) in a large meta-analysis, but not in single studies (2,27). Further powerful studies are expected to ascertain its association with CAD.
FUTURE PERSPECTIVES
CADgene is our first gene database for cardiovascular diseases aiming to provide a complete and up-to-date gene resource to the research community. We will continue collecting CAD-related association and GWAS data regularly. On the other hand, since there are many overlaps on the definitions and pathogeneses of some cardiovascular diseases, especially for CAD, MI and atherosclerosis, we plan to construct a comprehensive gene database for the main cardiovascular diseases and develop more tools to utilize the data conveniently in the future.
AVAILABILITY
The CADgene database is freely available at http://www.bioguo.org/CADgene/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Starting Fund from Huazhong University of Science and Technology (to A.Y.G.); Fundamental Research Funds for the Central Universities (2010MS045 to A.Y.G. and 2010MS015); China National 863 Scientific Program (2006AA02Z476); China National Basic Research Programs (973 Programs 2007CB512000); Key Academic Program Leader Award of Wuhan City (200951830560); Hubei Province Natural Science Key Program (2008CDA047); China National Natural Science Foundation (NSF30670857, 30800457). Funding for open access charge: China National Basic Research Programs (973 Programs 2007CB512000).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Chengqi Xu, Cong Li, Fan Wang, Dan Wang, Shaofang Nie, Sisi Li for their valuable advice and discussions.
REFERENCES
- 1.Kullo IJ, Ding K. Mechanisms of disease: the genetic basis of coronary heart disease. Nat. Clin. Pract. Cardiovasc. Med. 2007;4:558–569. doi: 10.1038/ncpcardio0982. [DOI] [PubMed] [Google Scholar]
- 2.Franchini M, Peyvandi F, Mannucci PM. The genetic basis of coronary artery disease: from candidate genes to whole genome analysis. Trends Cardiovasc. Med. 2008;18:157–162. doi: 10.1016/j.tcm.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Girelli D, Martinelli N, Peyvandi F, Olivieri O. Genetic architecture of coronary artery disease in the genome-wide era: implications for the emerging “golden dozen” loci. Semin. Thromb. Hemost. 2009;35:671–682. doi: 10.1055/s-0029-1242721. [DOI] [PubMed] [Google Scholar]
- 4.Fishbein MC. The vulnerable and unstable atherosclerotic plaque. Cardiovasc. Pathol. 2010;19:6–11. doi: 10.1016/j.carpath.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Kume N. Molecular mechanisms of coronary atherosclerotic plaque formation and rupture. Nippon. Rinsho. 2010;68:637–641. [PubMed] [Google Scholar]
- 6.Bui QT, Prempeh M, Wilensky RL. Atherosclerotic plaque development. Int. J. Biochem. Cell Biol. 2009;41:2109–2113. doi: 10.1016/j.biocel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q. Molecular genetics of coronary artery disease. Curr. Opin. Cardiol. 2005;20:182–188. doi: 10.1097/01.hco.0000160373.77190.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandona S, Stewart AF, Roberts R. Genomics in coronary artery disease: past, present and future. Can. J. Cardiol. 2010;26(Suppl. A):56A–59A. doi: 10.1016/s0828-282x(10)71064-3. [DOI] [PubMed] [Google Scholar]
- 9.Damani SB, Topol EJ. Future use of genomics in coronary artery disease. J. Am. Coll Cardiol. 2007;50:1933–1940. doi: 10.1016/j.jacc.2007.07.062. [DOI] [PubMed] [Google Scholar]
- 10.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Lu M, Shi L, Rui W, Zhu X, Chen G, Shang T, Tang J. Cardio: a web-based knowledge resource of genes and proteins related to cardiovascular disease. Int. J. Cardiol. 2004;97:245–249. doi: 10.1016/j.ijcard.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Bober M, Wiehe K, Yung C, Onal Suzek T, Lin M, Baumgartner W, Jr, Winslow R. CaGE: cardiac gene expression knowledgebase. Bioinformatics. 2002;18:1013–1014. doi: 10.1093/bioinformatics/18.7.1013. [DOI] [PubMed] [Google Scholar]
- 14.Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26:145–146. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanhoutte PM. Endothelial dysfunction: the first step toward coronary arteriosclerosis. Circ. J. 2009;73:595–601. doi: 10.1253/circj.cj-08-1169. [DOI] [PubMed] [Google Scholar]
- 18.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 19.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross R. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 22.Auer J, Weber T, Berent R, Lassnig E, Lamm G, Eber B. Genetic polymorphisms in cytokine and adhesion molecule genes in coronary artery disease. Am. J. Pharmacogenomics. 2003;3:317–328. doi: 10.2165/00129785-200303050-00003. [DOI] [PubMed] [Google Scholar]
- 23.Auro K, Alanne M, Kristiansson K, Silander K, Kuulasmaa K, Salomaa V, Peltonen L, Perola M. Combined effects of thrombosis pathway gene variants predict cardiovascular events. PLoS Genet. 2007;3:e120. doi: 10.1371/journal.pgen.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li AC, Palinski W. Peroxisome proliferator-activated receptors: how their effects on macrophages can lead to the development of a new drug therapy against atherosclerosis. Annu. Rev. Pharmacol. Toxicol. 2006;46:1–39. doi: 10.1146/annurev.pharmtox.46.120604.141247. [DOI] [PubMed] [Google Scholar]
- 25.Rajesh M, Mukhopadhyay P, Hasko G, Liaudet L, Mackie K, Pacher P. Cannabinoid-1 receptor activation induces reactive oxygen species-dependent and -independent mitogen-activated protein kinase activation and cell death in human coronary artery endothelial cells. Br. J. Pharmacol. 2010;160:688–700. doi: 10.1111/j.1476-5381.2010.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper PC, Rezende SM. An overview of methods for detection of factor V Leiden and the prothrombin G20210A mutations. Int. J. Lab. Hematol. 2007;29:153–162. doi: 10.1111/j.1751-553X.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- 27.Ye Z, Liu EH, Higgins JP, Keavney BD, Lowe GD, Collins R, Danesh J. Seven haemostatic gene polymorphisms in coronary disease: meta-analysis of 66,155 cases and 91,307 controls. Lancet. 2006;367:651–658. doi: 10.1016/S0140-6736(06)68263-9. [DOI] [PubMed] [Google Scholar]