Abstract
MicroRNAs (miRNAs), i.e. small non-coding RNA molecules (∼22 nt), can bind to one or more target sites on a gene transcript to negatively regulate protein expression, subsequently controlling many cellular mechanisms. A current and curated collection of miRNA–target interactions (MTIs) with experimental support is essential to thoroughly elucidating miRNA functions under different conditions and in different species. As a database, miRTarBase has accumulated more than 3500 MTIs by manually surveying pertinent literature after data mining of the text systematically to filter research articles related to functional studies of miRNAs. Generally, the collected MTIs are validated experimentally by reporter assays, western blot, or microarray experiments with overexpression or knockdown of miRNAs. miRTarBase curates 3576 experimentally verified MTIs between 657 miRNAs and 2297 target genes among 17 species. miRTarBase contains the largest amount of validated MTIs by comparing with other similar, previously developed databases. The MTIs collected in the miRTarBase can also provide a large amount of positive samples to develop computational methods capable of identifying miRNA–target interactions. miRTarBase is now available on http://miRTarBase.mbc.nctu.edu.tw/, and is updated frequently by continuously surveying research articles.
INTRODUCTION
As small non-coding RNAs of ∼22 nt, microRNAs (miRNAs) regulate gene expression post-transcriptionally through suppressing mRNA translation or inducing mRNA degradation by hybridizing to the 3′-untranslated regions (3′-UTR) of mRNAs. Discovery of the first miRNA in Caenorhabditis elegans in 1993 (1) ushered in numerous studies on the cellular processes of these tiny regulatory RNAs for a large variety of metazoan. Identified in mammalian cells over the past two decades in thousands of varieties, miRNAs play critical roles in many biological processes, including cell-cycle control, cell growth and differentiation, apoptosis, as well as embryo development.
Many miRNA-related database systems have been developed in recent years to provide further insight into miRNAs and their target genes. miRBase (2) is the most complete repository for miRNA annotation and nomenclature. Until now, the miRBase (version 16.0) contains 15 172 miRNA entries and many more new sequences are added regularly. miRGen (3), miRGator (4), miRDB (5), microRNA.org (6) and miRNAMap (7,8) provide miRNA targets by integrating extensively adopted target prediction programs. Moreover, TarBase (9), miRecords (10) and miR2Disease (11) contain experimentally validated miRNA–target interactions (MTIs). TarBase is the first resource to provide experimentally verified MTIs by surveying pertinent literature (9). miRecords accumulates both experimentally validated miRNA targets and computationally predicted miRNA targets (10). miR2Disease contains relationships among miRNAs, target genes and diseases in humans (11). miRSel (12) incorporates a text-mining method to extract systematically miRNA–target relationships from the PubMed abstracts. Additionally, among the several computational methods and web-based programs developed to identify target genes of miRNAs include miRanda (6), TargetScan (13), RNAhybrid (14), Pictar (15) and PITA (16). These extensively adopted research tools are highly promising for identifying MTIs, with their effectiveness confirmed experimentally.
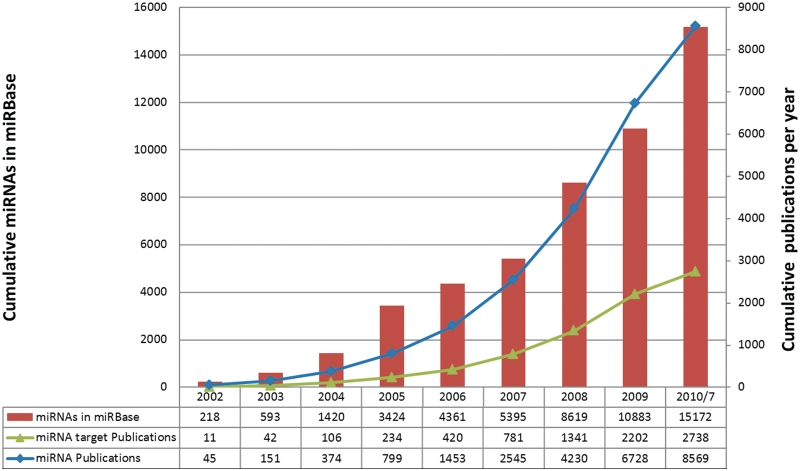
miRNA-related research has grown exponentially in recent years (Figure 1). The accelerated rate of miRNA gene discovery warrants a thorough investigation of the functions of these miRNAs. Additionally, more than 20 databases and computational methods have been developed to identify candidates of MTIs. A curated collection of updated MTIs with experimental support is essential to elucidating miRNA functions under different conditions and in different species. This work presents a database, miRTarBase, which has accumulated more than 3000 MTIs collected by manually surveying pertinent literature after adopting a systematic text-mining procedure to select research articles related to functional studies of miRNAs. Generally, the collected MTIs are validated experimentally based on reporter assays, western blot, or microarray experiments with overexpression or knockdown of miRNAs.
Figure 1.
Growth of miRNA genes in the miRBase database and the growth of the keywords with ‘miRNA’ and keyword with ‘miRNA target’ in PubMed.
Generally, an experimentally validated MTI initially involves using computational methods to identify target sites of miRNAs. These putative MTIs are then validated by molecular experiments, including reporter assays and western blot. Reporter assays and western blot are the conventional means of confirming the interaction between miRNA and its target mRNA. Additionally, northern blot analysis, quantitative real-time PCR (qPCR), or in situ hybridization is often performed to examine the co-expression of predicted miRNA and mRNA target gene. In contrast with conventionally adopted traditional validation approaches, genome-wide screening approaches have been developed, including microarray experiments with overexpression or knockdown of miRNAs, stable isotope labeling with amino acids in culture (SILAC) or pulsed SILAC (pSILAC). For instance, Selbach et al. determined the complement of all genes targeted by five miRNAs induced independently in HeLa cells using microarrays and pSILAC (17). That study identified more than 400 MTIs as well.
A growing number of miRNAs and their targets have been identified in recent years, combined with their major roles in biological systems, explaining the need to easily access an accurate, updated and centralized information repository. This work devises a frequently updated database by surveying continuously research articles with the pre-screening based on text-mining programs and the intention to make the database become a major repository for experimentally confirmed MTIs. In addition to containing the largest number of validated MTIs, the miRTarBase provides the most current collection by comparing with other similar databases developed previously, including TarBase, miRecords and miR2Disease. This work also elucidates the biological features of miRNA/target duplex based on more than 700 validated MTIs in human, where the miRNA target sites of MTIs have been described in previous literature. The MTIs collection in the miRTarBase can also become a largest amount of positive samples to develop computational methods in order to identify MTIs.
DATABASE CONTENT
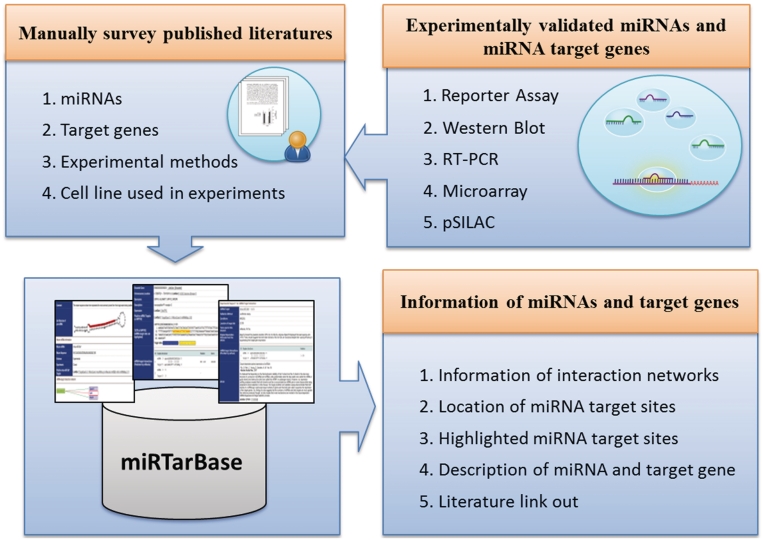
All database entries are collected manually to describe how a miRNA and its target genes are related to experimental support (Figure 2). Initially, all fields in the PubMed database are searched based on the keywords ‘microRNA targets’ or ‘miRNA targets’, followed by downloading the full text of these articles. A text-mining system is then devised to survey full-text literature that potentially describes MTIs, as verified by various experimental methods. Each research article is carefully reviewed by at least two of our developers to extract the MTIs, which are experimentally confirmed by reporter assays, western blot, microarray experiments, pSILAC or qRT–PCR. Additionally, other effective information is extracted, e.g. the species of miRNAs, species of target genes and experimental conditions.
Figure 2.
System flow of miRTarBase.
Strong experimental evidence of MTIs
The most effective means of verifying MTIs involves using fluorescence quantitative PCR and western blot methods to detect mRNA expression levels and protein expression levels under conditions of miRNA overexpression or miRNA knock-down cells. Despite the ability of the above methods to identify miRNA target genes accurately, other experimental methods must locate the regions targeted by miRNAs. Luciferase reporter assay is extensively adopted. Here, the MTIs are viewed as having strong support when they are validated by western blot, qPCR, or reporter assays.
Less strong experimental evidence of MTIs
High-throughput miRNA target identification methods, including pSILAC and microarray experiments, can determine the mRNA expression levels or protein expression levels when the miRNA is present or not (9). Given our inability to understand whether the over-expressed miRNAs cause the changed expression patterns directly or not, these technologies provide less strong experimental evidence of the collected MTIs.
Statistics and data analysis
In the release 1.0 (15 October 2010) of miRTarBase, 3576 curated MTIs between 657 miRNAs and 2297 target genes were collected from 985 articles. Table 1 lists the number of the collected MTIs in each species. For instance, 2531 human MTIs were collected between 287 miRNAs and 1631 target genes with the experimental support from 773 articles; in addition, 617 and 1409 interactions were confirmed experimentally by western blot and reporter assays, respectively. Each human miRNA can target five target genes on average. Supplementary Figure S1 shows the distribution of miRNAs categorized by the number of target genes for each miRNA which are supported by reporter assays or western blot. In the miRTarBase, hsa-miR-122 was recorded as having 45 target genes, which were validated experimentally by luciferase reporter assays or western blot. Notably, hsa-miR-122 is a liver-specific miRNA in humans and is significantly down-regulated in liver cancer (18).
Table 1.
Statistics of MTIs collected in miRTarBase
| Species | No. of MTIs | No. of miRNAs | No. of target genes | No. of articles collecteda | No. of MTIs experimentally validated by |
|||
|---|---|---|---|---|---|---|---|---|
| Strong evidences |
Less strong evidences |
|||||||
| Western blot | Reporter assays | pSILAC | Microarray | |||||
| Human | 2531 | 287 | 1631 | 773 | 617 | 1409 | 438 | 640 |
| Mouse | 336 | 110 | 236 | 141 | 172 | 269 | 0 | 40 |
| Rat | 232 | 95 | 92 | 40 | 77 | 43 | 0 | 159 |
| Chicken | 37 | 4 | 35 | 4 | 0 | 37 | 0 | 0 |
| Sheep | 5 | 5 | 2 | 2 | 0 | 2 | 0 | 0 |
| Zebrafish | 82 | 27 | 62 | 23 | 14 | 75 | 0 | 0 |
| Fruit fly | 128 | 39 | 80 | 31 | 9 | 126 | 0 | 0 |
| Silkworm | 2 | 2 | 1 | 1 | 0 | 2 | 0 | 0 |
| Nematode | 32 | 6 | 27 | 20 | 4 | 27 | 0 | 0 |
| Plants | 172 | 69 | 121 | 11 | 7 | 2 | 0 | 22 |
| Viruses | 19 | 13 | 10 | 8 | 1 | 19 | 0 | 0 |
| Total | 3576 | 657 | 2297 | 1054 | 901 | 2011 | 438 | 861 |
aArticles may report various MTIs in different species.
This work also examines the functions of these target genes involved in human MTIs collected in the database by performing gene ontology (GO) and KEGG (19) pathway enrichment annotation using the DAVID gene annotation scheme (20). GO enrichment analysis indicates that the cellular process, biological regulation and metabolic process are the most significantly enriched GO terms to select human target genes (Supplementary Figure S2). Supplementary Table S1 lists the top 20 pathways significantly enriched in these human target genes, most of which are involved in cancer, including prostate, pancreatic, colorectal, small cell lung, endometrial, non-small cell lung and bladder. Interestingly, above analysis provides an overview of the possible functions of human miRNAs based on this curation of MTIs. This is despite the fact that the data should be biased since miRNAs have attracted increasing attention in cancer research recently.
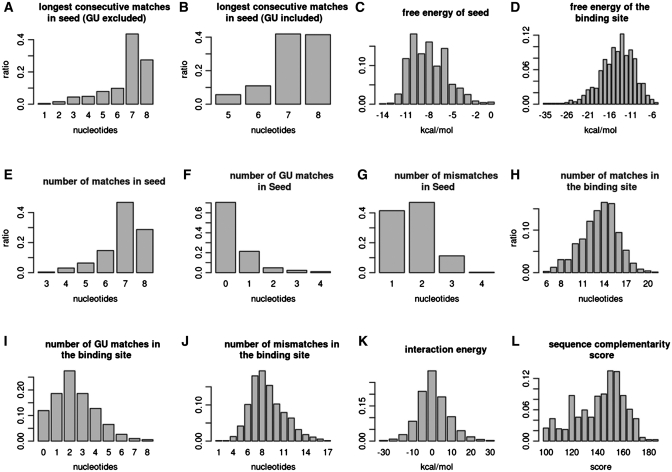
Only the 709 human MTIs in the miRTarBase have miRNA target site annotations, which can be extracted from the articles. Of these target site sequences, nine of them only provide the sequence of seed region (<10 nt); 667 of them contain the target site sequences (10–50 nt), while the remaining ones (33) provide cloned partial UTR sequences (>50 nt). Next, an attempt is made to summarize the data distributions of 12 biological features of the miRNA/target duplex in these 709 known human MTIs, as shown in Figure 3. The miRNA target sites are mapped to the 3′-UTR of the corresponding target gene; in addition, 70 nt around the target site are extracted. Moreover, the miRNA target sites are selected when the alignment score (by mirnada) of miRNA/target duplex exceeds 100 and the number of base pairs within the seed region is higher than5. Notably, 721 miRNA target sites are obtained from the 709 MTIs. Figure 3 displays the histograms of various features of these miRNA–target duplexes. Figure 3A and B show the longest consecutive matches (excluding or including wobble pairing (GU pairing) in a seed region), i.e. a subsequence from nucleotide 1–8 in the 5′-end of the miRNA, respectively. More than 80% of all binding sites have more than seven bases of consecutive pairings. The minimum free energy of the seed regions and the binding sites is also calculated, as shown in Figure 3C and D, respectively. The mean value of the free energy of the binding site is approximately −14 kcal/mol. The free energy of most of the seeds is lower than −6 kcal/mol. Next, analysis is performed of the number of nucleotides matches, GU matches, and mismatches in the seed regions and the target sites. Figure 3E and J summarize these statistics. More than 85% of all target sites have at least six matches in the seed region; in addition, the GU matches rarely occur in the seed region, i.e. <30%. The number of matches is significantly higher than the number of mismatches. GU matches in the target sites are significantly smaller than the quantity of matches and mismatches. The interaction energy is also estimated based on the calculations of Kertesz M. et al. (16). According to our results, most of the interaction energy of target site accessibility shown in Figure 3K ranges from −10 to 10 kcal/mol.
Figure 3.
Histograms of various features of an experimentally confirmed miRNA/target duplex.
Comparison with other MTI databases
Comparing the other manually curated databases such as TarBase, miRecords, and miR2Disease reveals that miRTarBase accumulates a larger collection and a more updated curation of MTIs than other resources (Table 2). In particular, around 900 research articles are collected in our database. This comparison also reveals that the proposed miRTarBase has the most abundant MTIs, even if only considering the entries supported by reporter assays or western blot experiments. Furthermore, the Venn diagrams display the intersection of articles collected in different databases (Supplementary Figure S4). miRTarBase covers all of the research articles collected in TarBase, miRecords and miR2Disease.
Table 2.
Comparison of miRTarBase with other MTI databases
| Database names | TarBase (9) | miRecords (10) | miR2Disease (11) | miRTarBase |
|---|---|---|---|---|
| Support species | Metazoa × 6 | Metazoa × 9 | Human | Metazoa × 9 |
| Viridiplantae | Viruses × 2 | Viridiplantae × 3 | ||
| Viruses | Viruses × 5 | |||
| Total No. of miRNAs | 223 | 381 | 179 | 657 |
| Total No. of target genes | 1028 | 1057 | 394 | 2297 |
| Total No. of articles | 154 | 410 | 421 | 985 |
| Total No. of MTIs | 1264 | 1513 | 635 | 3576 |
| Supported by strong experimental evidences | ||||
| No. of MTIs validated by ‘Reporter assays’ | 305 | 672 | 635 | 2017 |
| No. of MTIs validated by ‘Western blot’ | 27 | 295 | 0 | 901 |
| No. of MTIs validated by ‘Reporter assays and western blot’ | 25 | 123 | 0 | 711 |
| No. of MTIs validated by ‘Reporter assays or western blot’ | 307 | 747 | 635 | 2207 |
| Supported by less strong experimental evidences | ||||
| No. of MTIs validated by ‘pSILAC experiments’ | 455 | 0 | 0 | 455 |
| No. of MTIs validated by ‘Microarray experiments’ | 343 | 380 | 0 | 861 |
Alternatively, the text-mining method can retrieve the information of relations between miRNAs and target genes. However, MTI is generally described in a natural language and is not easily extracted correctly by only computational methods. For instance, although important genes involved in the biogenesis of miRNA, DICER1 and Drosha are normally not the target genes of a miRNA when they are discussed along with the miRNA in an article. However, text-mining methods may identify the relation between a miRNA and DICER1, and incorrectly annotate the relation as a MTI. Therefore, manually reviewing the articles that may contain MTIs is inevitable for extracting such experimental evidence to support a MTI. Notably, this study does not compare the contents of the miRTarBase with those of other databases established by only text-mining methods without manual review.
WEB INTERFACE
miRTarBase provides various query interfaces and graphical visualization pages to facilitate the access of MTI data (Supplementary Figure S3). Several search functions for retrieving MTIs are designed, including search by miRNA accessions, search by target genes, and search by literature. Alternatively, miRTarBase provides a keyword search in all fields for all data entries. Here, a result page is designed to describe MTI, where each MTI is assigned a miRTarBase accession. The result page largely comprises three main parts, i.e. miRNA information, target gene information and evidence support. Generally, web pages of the miRTarBase contain many effective quick links to several other web resources, including NCBI Entrez (21), UCSC Genome Browser (22), miRBase (2), BioGPS (23), iHOP (24) and HGNC (25). The web pages are described in detail below.
The ‘miRNA information’ page contains the attributes of a miRNA such as accession, synonyms, descriptions, miRNA sequence, and links to other putative MTI databases. In particular, in this page all MTIs of miRNA are presented as a network, which can depict the relationships between a miRNA and multiple target genes. In the ‘Target Gene’ page, the basic information of a target gene is provided, including a gene symbol, description, genomic location, transcript sequence and links to other resources. Target site-related information located in the transcript is carefully examined and displayed on the web page. Notably, many articles only describe the regulatory relationship between a miRNA and its target genes without providing the exact regions of miRNA target sites. By using miRanda, this work attempts to identify computationally the potential target sites belonging to a MTI, which is supported by experimental evidence.
In the ‘Evidence’ page, the experimental information to support a MTI from one or multiple articles is provided by presenting the experimental validation methods, experimental conditions, location of target sites, computational tools used in an article, partial key descriptions extracted from the article and article abstract. Additionally, this resource also provides a data submission page that allows users or researchers to submit information of MTIs, which have not yet been curated. The database provides a convenient approach for users to directly suggest articles containing information about MTI, followed by review of the suggested articles by the developer of miRTarBase.
SUMMARY AND PROSPECTICTIVE WORKS
This work presents a more comprehensive collection of MTIs, which are validated experimentally. The biological features of miRNA/target duplex are observed based on the largest collection of human MTIs currently available. Various web interfaces are also designed to facilitate the presentation of MTIs. Moreover, a pipeline combining text-mining and manual review methods is established to extract MTI information from research articles.
Future work involving the proposed database should extend the human MTIs to mouse, rat and other mammalian genome based on evolutionary conservation of miRNA and its target sites. More probable MTIs can be provided as promising candidates for experimental confirmation.
AVAILABILITY
miRTarBase database will be continuously maintained and updated monthly. The database is now publically accessible through the URL http://miRTarBase.mbc.nctu.edu.tw/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council of the Republic of China (Contract No. NSC 98-2311-B-009-004-MY3 and NSC 99-2627-B-009-003); National Research Program For Genomic Medicine (NRPGM), Taiwan; MOE ATU (partial). Funding for open access charge: National Science Council, Taiwan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Ted Knoy is appreciated for his editorial assistance. Special thanks for the financially supports from National Research Program for Genomic Medicine (NRPGM), Taiwan.
REFERENCES
- 1.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexiou P, Vergoulis T, Gleditzsch M, Prekas G, Dalamagas T, Megraw M, Grosse I, Sellis T, Hatzigeorgiou AG. miRGen 2.0: a database of microRNA genomic information and regulation. Nucleic Acids Res. 2010;38:D137–D141. doi: 10.1093/nar/gkp888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36:D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naeem H, Kuffner R, Csaba G, Zimmer R. miRSel: Automated extraction of associations between microRNAs and genes from the biomedical literature. BMC Bioinformatics. 2010;11:135. doi: 10.1186/1471-2105-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 16.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 17.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 18.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 19.Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez JM, Hoffmann R, Valencia A. iHOP web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]