Abstract
Peroxisome proliferator-activated receptor α (PPARα) is a key regulator of lipid homeostasis in hepatocytes and target for fatty acids and hypolipidemic drugs. How these signaling molecules reach the nuclear receptor is not known; however, similarities in ligand specificity suggest the liver fatty acid binding protein (L-FABP) as a possible candidate. In localization studies using laser-scanning microscopy, we show that L-FABP and PPARα colocalize in the nucleus of mouse primary hepatocytes. Furthermore, we demonstrate by pull-down assay and immunocoprecipitation that L-FABP interacts directly with PPARα. In a cell biological approach with the aid of a mammalian two-hybrid system, we provide evidence that L-FABP interacts with PPARα and PPARγ but not with PPARβ and retinoid X receptor-α by protein–protein contacts. In addition, we demonstrate that the observed interaction of both proteins is independent of ligand binding. Final and quantitative proof for L-FABP mediation was obtained in transactivation assays upon incubation of transiently and stably transfected HepG2 cells with saturated, monounsaturated, and polyunsaturated fatty acids as well as with hypolipidemic drugs. With all ligands applied, we observed strict correlation of PPARα and PPARγ transactivation with intracellular concentrations of L-FABP. This correlation constitutes a nucleus-directed signaling by fatty acids and hypolipidemic drugs where L-FABP acts as a cytosolic gateway for these PPARα and PPARγ agonists. Thus, L-FABP and the respective PPARs could serve as targets for nutrients and drugs to affect expression of PPAR-sensitive genes.
Peroxisome proliferator-activated receptor α (PPARα) is a nuclear target for fatty acids, hypolipidemic drugs, and other peroxisome proliferators (1–4) and initiates gene expression of enzymes involved in lipid metabolism (5, 6). Two further subtypes of this receptor exist, namely PPARβ and PPARγ, of which the latter is implicated to play a role in adipogenesis and adipocyte fatty acid metabolism upon activation by fatty acids and antidiabetic thiazolidindiones (7). The mechanism and pathway by which fatty acids and respective drugs as signaling molecules reach their destination are not known, but assuming targeted transport in hepatocytes where all three subtypes of this receptor are expressed (8), liver fatty acid binding protein (L-FABP) is a candidate to serve as shuttle for these ligands. This hypothesis is based on several observations. First, the 14.4-kDa L-FABP, which is supposed to play a role in intracellular lipid trafficking (9, 10), is abundant in the cytosol and is also found inside the nucleus of liver cells (11), the presumed place of PPAR activation. Second, L-FABP binds fatty acids and hypolipidemic drugs that have been identified as PPARα agonists (2–4) as well as BRL48,482, an antidiabetic thiazolidindione, all with dissociation constants in the micro to nanomolar range (12–14). Third, hypolipidemic drugs are able to induce expression of L-FABP and β-oxidative enzymes via PPARα (15). This link between multiple ligand interactions and gene expression was revealed recently by us by identifying branched-chain phytanic acid as a ligand for L-FABP and PPARα and by demonstrating that this fatty acid induced L-FABP expression via activation of PPARα (6). Thus, L-FABP might be part of the PPARα targeted signal transduction pathway, for which two alternative mechanisms can be envisaged (10, 16); L-FABP forms a cytosolic sink for the signaling molecules, thus acting as negative regulator of their concentrations available for PPARα activation in the nucleus (i.e., increased PPARα activation results from decreased intracellular L-FABP concentrations). Alternatively, L-FABP itself transports the signaling molecules to the nucleus to activate PPARα, which implies a positive correlation between PPARα activation and intracellular L-FABP concentrations. These options provided us with a rationale for elucidating a clearly defined cellular task for L-FABP with regard to gene expression.
We hypothesized that L-FABP in the nucleus interacts with the PPAR isoforms and designed experiments that could provide evidence of a direct protein–protein interaction. Furthermore, the availability of established antisense L-FABP mRNA HepG2 cell lines with varying intracellular concentrations of L-FABP (17) enabled us to verify one of the two hypotheses mentioned above by using a cell culture model that endogenously expresses L-FABP in the context of a well preserved lipid metabolism.
Materials and Methods
Materials.
Phytanic acid and bezafibrate were obtained from Sigma; Wy14,643 and ciglitazon were from Biomol (Plymouth Meeting, PA). Oligonucleotides were purchased from MWG Biotec (Ebersberg, Germany), restriction enzymes were from Roche Diagnostics, and [35S]methionine (1 Ci/mol) was from Amersham Pharmacia. All chemicals used were of analytical grade.
Plasmids.
Expression plasmids for human and murine PPARα were obtained by cloning of the cDNA into the pCDNA3 mammalian expression vector (Promega); expression plasmids for human and murine PPARβ and PPARγ were kindly provided by Walter Wahli (University of Lausanne). Expression plasmid for murine RXRα was kindly provided by Pierre Chambon (Université Louis Pasteur, Strasbourg).
Transactivation Experiments.
For transactivation studies, ideal peroxisome proliferator-responsive element (PPRE) (18) was cloned into the pCAT3-promoter plasmid (Promega). HepG2 cells (American Type Culture Collection, HB-8065) and antisense L-FABP mRNA HepG2 cells, derived after stable transfection with the complete cDNA of human L-FABP in antisense orientation (17), were grown to 60–70% confluency in 6-well dishes (Nunc) in DMEM (Biochrom, Berlin) supplemented with 10% Basal-Medium-Supplement artificial serum, fatty acid free (Biochrom), and then transfected with the reporter gene (1.5 μg) by using pSV-β-galactosidase (β-Gal) (Promega) as internal reference (0.5 μg) and cotransfected with murine PPARα or PPARγ-expressing plasmid with the aid of Fugene6 (Roche Diagnostics). After transfection, cells were incubated for 24 h with the respective ligand, with concentrations ranging from 50 to 200 μM (19, 20). DMSO concentration was kept below 1%. Chloramphenicol acetyltransferase (CAT) and β-Gal expressions were measured by using an ELISA system (Roche Diagnostics). L-FABP was determined by an established ELISA system (17).
In Vitro Pull-Down Assay.
PPARα was translated in vitro by using the TNT kit (Promega) with [35S]methionine according to the manufacturer's description. L-FABP–Sepharose was prepared by covalently binding 10 mg of recombinant L-FABP to CH-activated Sepharose (Amersham Pharmacia). Radiolabeled protein purity was checked by SDS/PAGE and subsequently incubated with L-FABP–Sepharose for 1 h at 4°C. After washing of the precipitate, the proteins were eluted with SDS-loading buffer and separated by SDS/PAGE. 35S-radiolabeled PPARα was detected by autoradiography.
To assess whether L-FABP–PPAR interaction is ligand dependent, we modified the in vitro pull-down assay. First, we determined the concentration of binding sites on delipidated L-FABP–Sepharose to be 5 ± 1 μM (n = 5) by titration with [1-14C]oleic acid. The assay (n = 6, DMSO as ligand solvent was always kept at 1% in the assay) then was carried out with no ligand present (DMSO only) on the one hand, and after incubation with 100 μM (i.e., 20-fold excess of binding sites available), of either linoleic acid or Wy14,643 on the other hand. Loading of delipidated L-FABP–Sepharose was carried out for 15 min at room temperature, then temperature was lowered to 4°C to freeze equilibrium; the experiment was then carried out as described above. Binding of [35S]PPARα was quantified by liquid scintillation counting; purple acid phosphatase-Sepharose served as negative control.
Immunocoprecipitation.
Anti-L-FABP–Sepharose was prepared by covalently binding 20 mg of polyclonal rabbit anti-murine L-FABP antibody (6) to CH-activated Sepharose. Nuclear lysates of mouse liver were prepared according to Stümpfle et al. (21) from 5 g of tissue. The nuclear fraction obtained was incubated with Sepharose-bound antibodies for 1 h at 4°C. After washing of the precipitate, the proteins were eluted with SDS-loading buffer, separated by SDS/PAGE (13.5%), and detected by Western blotting with (i) rabbit anti-murine L-FABP antibody (7.5 μg/ml) and anti-rabbit IgG-horseradish peroxidase antibody (1:8,000) (Sigma), and (ii) goat anti-PPARα (10 μg/ml) (Santa Cruz Biotechnology) and anti-goat IgG-horseradish peroxidase antibody (1:10,000) (Sigma). Proteins were visualized by chemiluminescence detection by using the ECL system (Amersham Pharmacia).
Mammalian Two-Hybrid Assay.
For use of the mammalian two-hybrid assay system (CLONTECH), COS7 cells (American Type Culture Collection) were grown to 60–70% confluency in 6-well dishes (Nunc) in basal Iskov's modified Eagle's medium (Biochrom) supplemented with 8% FCS and transfected by using Fugen6 (Roche Diagnostics) with 1 μg of expression vector of a fusion protein of the GAL4-DNA binding domain to PPARα, PPARβ, PPARγ, or RXRα (GAL4DBD-PPAR/RXR, bait protein) and 1 μg of a fusion protein of the VP16 activation domain to L-FABP (VP16AD-L-FABP, target protein). In addition, COS7 cells were transfected for 48 h with 1 μg of CAT-reporter gene vector under the control of a GAL4-responsive element and 0.1 μg of β-Gal normalization vector. CAT and β-Gal expression were quantified by ELISA (Roche Diagnostics). To measure unspecific interaction expression, vectors for bait and target protein were changed to expression vectors for a fusion protein of GAL4-DNA binding domain to p53 (GAL4DBD-p53) or to a fusion protein of VP16 activation domain to the simian virus 40-T-antigen (VP16AD-SV40T). For positive and negative controls, respective expression vectors supplied with the kit were used. To assess the effect of ligand on L-FABP–PPARα interaction, cells were treated after transfection for 48 h with 100 μM linoleic acid and Wy14,643, respectively.
Results
Transactivation of Human PPARα by Fatty Acids and Hypolipidemic Drugs.
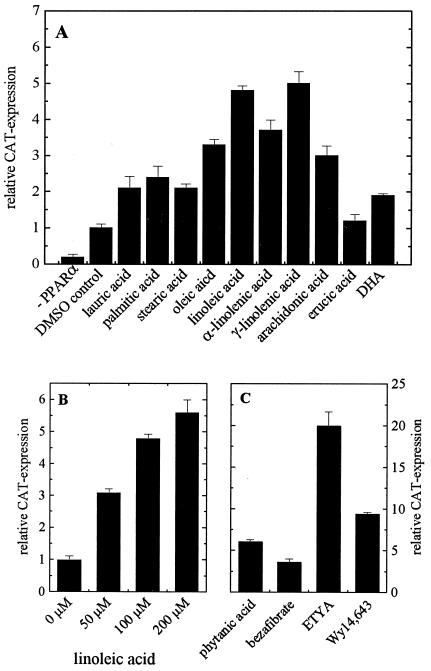
To verify our cell culture model with respect to PPARα activation, we transfected the human hepatoma HepG2 cell line with a PPARα-sensitive CAT-reporter gene carrying the ideal PPRE (18) together with the expression vector for human PPARα. The latter transfection became necessary because natural PPAR concentrations in the cells are too low for furnishing statistically meaningful transactivation data (Fig. 1A). Ligands were then applied for 24 h to the cells (4), which were lysed afterward; transactivation potential of the respective ligand was determined by measuring CAT expression relative to β-Gal expression via respective ELISAs. In general, unsaturated fatty acids are more potent agonists than saturated fatty acids (Fig. 1A), with the exception of monounsaturated erucic acid, which does not affect PPARα activation at all. By applying fatty acid concentrations up to the highest amount tolerated by the HepG2 cells, we show here that PPARα activation is concentration dependent (Fig. 1B). Branched-chain phytanic acid, the natural peroxisome proliferator and activator of murine PPARα (4), activates human PPARα comparable with that of hypolipidemic bezafibrate and Wy14,643, but about two times lower than eicosatetraynoic acid (ETYA), an analog of arachidonic acid (Fig. 1C).
Figure 1.
Transactivation of human PPARα in HepG2 cells. HepG2 cells were transfected with the expression vector for human PPARα, pSV-β-Gal, and the CAT-reporter gene vector under the control of ideal PPRE. (A) Cells were treated for 24 h with 100 μM fatty acid, except 50 μM for arachidonic acid and docosahexaenoic acid. As control, cells were either transfected with pCDNA3 instead of the expression vector for human PPARα (−PPARα) or treated with DMSO alone. (B) Concentration-dependent transactivation by linoleic acid (results are representative for all fatty acids tested). (C) Cells were treated for 24 h with phytanic acid (100 μM), bezafibrate (100 μM), ETYA (50 μM), and Wy14,643 (100 μM). β-Gal and CAT concentrations were determined by ELISAs; DMSO control was set as one. Each value represents the mean of six independent experiments ± SD.
In Vitro Tests for Nuclear Interaction of Murine L-FABP and PPARα.
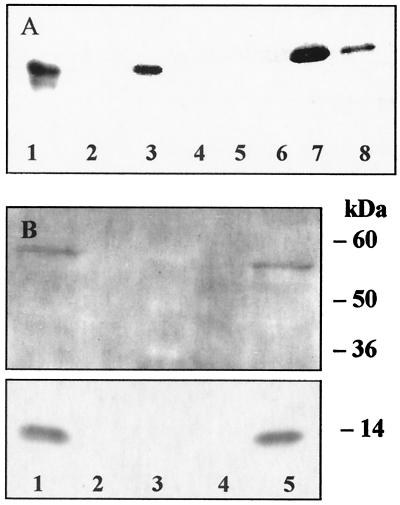
Laser-scanning microscopy of immunofluorescent-labeled L-FABP and PPARα, respectively, revealed colocalization of the two proteins in nuclei of mouse primary hepatocytes (data not shown) and agreed with earlier findings on the nuclear localization of L-FABP (11). This finding suggested to us that L-FABP functions as a carrier for PPARα agonists and might directly interact with the nuclear receptor. To verify this hypothesis, we set up first an in vitro binding experiment by incubating L-FABP immobilized on CH-activated Sepharose with 35S-radiolabeled PPARα. Complex formation was proven after removal from Sepharose and concomitant separation of the complex by SDS loading buffer, followed by subsequent analysis of radiolabeled PPARα by SDS/PAGE and autoradiographic detection (Fig. 2A, lane 7). In negative controls, 35S-labeled PPARα was incubated with Sepharose alone (Fig. 2A, lane 4) or with purple acid phosphatase covalently linked to Sepharose (Fig. 2A, lane 2), which produced no interaction. Specific binding was tested by eluting bound 35S-labeled PPARα with free L-FABP, resulting in partial removal of the labeled protein from the complex (Fig. 2A, lane 8).
Figure 2.
Direct interaction of murine L-FABP and murine PPARα. (A) Pull-down assay. Murine 35S-labeled PPARα (lanes 1 and 3, positive control) was precipitated with murine L-FABP covalently bound to CH-activated Sepharose and centrifuged; no 35S-PPARα was found in the supernatant (lane 5). The wash with PBS is free of 35S-PPARα (lane 6); thereafter, bound 35S-PPARα was eluted with SDS-loading buffer (lane 7). To test specific binding, the precipitate with bound 35S-PPARα was washed with PBS/murine L-FABP solution and eluted as described above (lane 8). To check for unspecific binding, purple acid phosphatase covalently linked to Sepharose (lane 2) or unmodified Sepharose (lane 4) was incubated with 35S-PPARα. Protein fractions obtained were separated by SDS/PAGE (13.5%), and 35S-PPARα was visualized by autoradiography. (B) Immunocoprecipitation (Upper, stained for PPARα; Lower, stained for L-FABP). From nuclear lysates of mouse liver (lane 1, positive control), L-FABP–PPARα complex was precipitated with anti-murine L-FABP antibody immobilized on Sepharose. Neither L-FABP nor PPARα was found in the supernatant (lane 4). After washing the precipitate with PBS, L-FABP and PPARα were eluted with SDS-loading buffer (lane 5). For negative control, unmodified Sepharose was used (lane 2). Protein fractions obtained were separated by SDS/PAGE (13.5%), and bands were visualized after Western blotting and immunodecoration; protein size was determined by molecular mass marker (lane 3).
To assess whether ligands modulate L-FABP–PPARα interaction, an assay similar to that described above was designed that quantified L-FABP–PPARα interaction by measuring the amount of radioactive PPARα bound to L-FABP–Sepharose in the absence and presence of ligand. L-FABP–Sepharose was first used in delipidated form where binding of 35S-labeled PPARα amounted to (2.52 ± 0.11) × 106 cpm. The value for 35S-labeled PPARα binding to L-FABP–Sepharose, when the latter was loaded to saturation with linoleic acid, was (2.43 ± 0.12) × 106 cpm, with Wy14,643 (2.49 ± 0.12) × 106 cpm. Unspecific binding (purple acid phosphatase-Sepharose) was only (0.45 ± 0.09) × 106 cpm. This experiment clearly demonstrated that L-FABP without ligand or loaded with either linoleic acid or Wy14,643 binds to PPARα with the same affinity. Thus, it is clear that the ligands do not affect L-FABP–PPARα interaction.
The approach with recombinant proteins was complemented with experiments designed to prove the interaction of the native proteins in lysates of mouse hepatocyte nuclei by immunocoprecipitation. Anti-L-FABP antibodies were immobilized on CH-activated Sepharose and applied to the nuclear lysate for 1 h. Upon precipitation, proteins bound were removed from the Sepharose by SDS loading buffer, separated by SDS/PAGE, and identified by Western blotting using specific antibodies for L-FABP and PPARα, respectively (Fig. 2B). The stains of native L-FABP and PPARα as shown in Fig. 2B, lane 5, clearly indicate complex formation in mouse liver. When nuclear lysate was incubated with Sepharose alone, neither L-FABP nor PPARα was bound (Fig. 2B, lane 2, negative control). Thus, it can be concluded that by direct protein–protein interaction, a complex of L-FABP and PPARα is formed in the nuclei of murine hepatocytes.
Verification of Protein–Protein Interaction of Murine Nuclear Receptors with L-FABP Using the Two-Hybrid Assay.
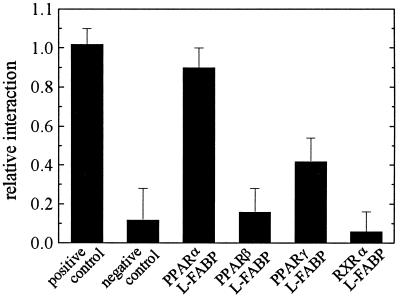
As in liver, where not only PPARα but also the β and γ subtypes are expressed, we wanted to test whether the interaction of L-FABP is restricted to the α subtype of the nuclear receptor. To this end, we used a mammalian cell-based two-hybrid assay and measured interaction of L-FABP with PPARα, PPARβ, and PPARγ as well as with RXRα (Fig. 3). Our in vitro data were borne out of the fact that interaction exists between L-FABP and PPARα in the same strong order of magnitude as that of the positive control supplied with the assay system. The interaction between L-FABP and PPARγ is about 3-fold weaker, and no interaction is observed between L-FABP and either PPARβ or RXRα. Again, to test whether ligands influence interaction of L-FABP with PPAR subtypes, we repeated the experiments by treating cells in addition with linoleic acid or Wy14,643. No change in CAT expression was observed (data not shown).
Figure 3.
Two-hybrid interaction of L-FABP with nuclear receptors. Interaction was measured by using the mammalian two-hybrid assay in COS7 cells, using a fusion protein of the GAL4-DNA binding domain with PPARα, PPARβ, PPARγ, or RXRα and a fusion protein of the VP16 activation domain with L-FABP. Unspecific interaction was quantified by using a fusion protein of the GAL4-DNA binding domain with p53 or a fusion protein of the VP16 activation domain with the simian virus 40-T antigen. Positive and negative controls were used according to the supplier's manual. CAT and β-Gal expression was measured by ELISAs. Each column represents the mean of 5–8 independent experiments ± SD.
Dependence of PPARα Transactivation on L-FABP Concentration in HepG2 Cells.
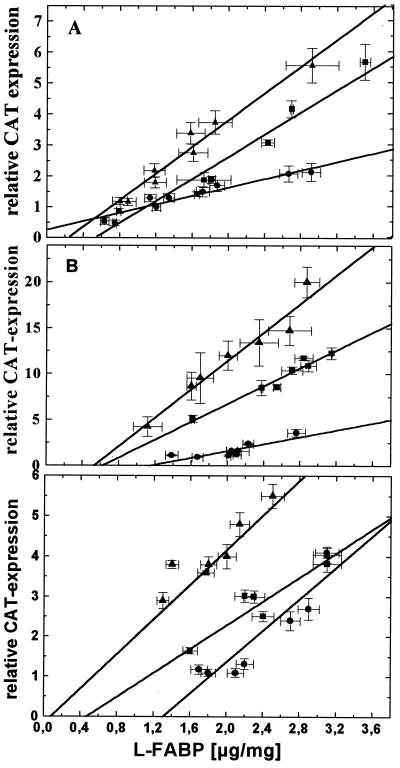
After this demonstration of direct L-FABP–PPARα and L-FABP–PPARγ interactions, we expected in our approach to derive quantitative data that L-FABP is a positive regulator of PPARα and PPARγ transactivation. A series of HepG2 cell clones, which express L-FABP at levels down to one-sixth of the normal after transfection with antisense L-FABP mRNA (17), was transfected with the ensemble of vectors consisting of a CAT-reporter gene vector under the control of the ideal PPRE (18), a β-Gal normalization vector, and an expression vector for either human PPARα or PPARγ and again incubated the cultures for 24 h with the series of fatty acids and hypolipidemic drugs. In addition to the determination of CAT and β-GAL expression by respective ELISAs, we measured intracellular L-FABP concentrations by ELISA (17). The clones investigated did not exhibit any changes in HepG2 growth characteristics. As shown in Fig. 4, the plots of transactivation (indicated by relative CAT expression) versus L-FABP concentration reveal linear relationships with positive slopes for all ligands tested, i.e., the less L-FABP the less PPARα transactivation. Of all fatty acids applied to HepG2 cells, phytanic acid affords the steepest and stearic acid the shallowest slope, i.e., 6.1-fold and 2-fold transactivation of PPARα, respectively (Fig. 4A). Not shown are the data for oleic, α-linolenic, and arachidonic acids, which also reveal a linear correlation between transactivation and L-FABP concentration, with values ranging between 4.6-fold for αlinolenic acid and 3.1-fold transactivation for arachidonic acid. The hypolipidemic drugs, except bezafibrate, are more potent PPARα activators than fatty acids (Fig. 4B). Extrapolation of all graphs shown in Fig. 4 to zero L-FABP concentration indicates that relative CAT expressions approach the value of one, i.e., no induction of transactivation. When no ligands were applied to the cells, the same correlation was found between L-FABP concentration and PPARα activation (data not shown). Also, extrapolation of the graph to zero L-FABP concentration led to an abolishment of PPARα activation. A linear correlation was also found in the case of PPARγ activation modulated by intracellular L-FABP content (Fig. 5C). Ciglitazon, a known potent activator of PPARγ, showed the highest PPARγ activation potential (5.5-fold), followed by Wy14,643 and linoleic acid with 4-fold and 3.8-fold activation potential, respectively. Also here, extrapolation of the graphs to zero L-FABP concentration revealed loss of PPARγ activation.
Figure 4.
Transactivation of human PPARα depends on L-FABP concentration. A series of eight HepG2 cell clones, with different L-FABP contents after stable transfection with antisense L-FABP mRNA (17), were transfected with the expression vector for human PPARα, pSV-β-Gal, and the CAT-reporter gene vector under the control of ideal PPRE. Each data point represents the analysis of a single clone with ELISAs for the determination of β-Gal, CAT, and L-FABP concentrations. (A) Cells treated for 24 h with 200 μM stearic acid (●), 200 μM linoleic acid (▴), and 100 μM phytanic acid (■). (B) Cells treated for 24 h with 200 μM bezafibrate (●), 50 μM ETYA (▴), and 200 μM Wy14,643 (■). (C) Cells treated for 24 h with 100 μM linoleic acid (●), 100 μM ciglitazon (▴), and 100 μM Wy14,643 (■). DMSO control was set as one. Note the different scale of ordinates. Each data point represents the mean of six independent experiments ± SD. P < 0.001 for all graphs in A and B; P < 0.004 for all graphs in C.
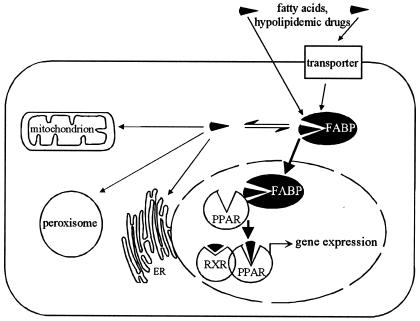
Figure 5.
Scheme for L-FABP action in PPAR-mediated gene regulation.
Discussion
The experiments carried out in this study provide evidence that a fatty acid binding protein is partner in gene regulation via PPARα and PPARγ in hepatocytes. Of the two mechanisms proposed, L-FABP definitely plays the role of positive regulator of PPARα and PPARγ activity, with nuclear receptor activity being strictly dependent on intracellular L-FABP concentrations. Furthermore, we show that murine L-FABP and PPARα as well as PPARγ interact via protein–protein contacts. Incidentally for FABPs in general, this is evidence for such contacts.
Our data on human PPARα transactivation are generally consistent with previously reported activation profiles of this nuclear receptor (1, 2, 4). ETYA is a strong activator of human PPARα, as shown by others (22) and by us in this study, thus validating the use of the HepG2 cell culture model. A further reason for choosing this model was the opportunity to study L-FABP function in cells that endogenously express this protein in the context of a well preserved lipid metabolism. It is interesting to note that phytanic acid, which we recognized earlier as a 6.2-fold activator of murine PPARα in HepG2 cells transfected with this nuclear receptor (4), has a similar stimulatory effect on human PPARα (6.1-fold). Thus, phytanic acid, at present, is one of the most potent naturally occurring activators of this isoform of PPARs.
Because L-FABP does not only bind fatty acids and hypolipidemic bezafibrate, ETYA and Wy14,643, and antidiabetic thiazolidindiones but also long-chain acyl-CoAs (13, 23), the question arises whether these fatty acid metabolites also might be ligands for PPARα. Literature reports reveal, however, that long-chain acyl-CoAs in contrast to fatty acids are not able to induce complex formation of PPRE and PPARα/RXRα, and inhibition of long-chain acyl-CoA synthetase with triascin C results in an increase in PPARα activation in vitro (2). Moreover, cotransfection of cells with a reporter plasmid preceded by the PPRE containing acyl-CoA oxidase promoter with expression plasmids for PPARα and long-chain acyl-CoA synthetase inhibited PPARα-mediated transactivation (24). Because we demonstrated earlier that fatty acid binding to PPARα can be correlated with its activation capability (4), we conclude that it is the fatty acid that directly affects PPARα in our studies (Fig. 5).
PPARα controls peroxisomal acyl-CoA oxidase (25) and bifunctional enzyme (26), small-, medium- and long-chain acyl-CoA dehydrogenases, and the trifunctional enzyme (27) of mitochondrial β-oxidation. Furthermore, PPARα controls regulation of genes encoding lipoprotein lipase and different apolipoproteins (27–29). Thus, PPARα can be considered a cellular sensor for fatty acids that controls their degradation and storage by stimulating gene expression of enzymatic and nonenzymatic proteins involved in their metabolism. As evidenced in this work, PPARα-mediated gene expression is also regulated by L-FABP, which controls the flux of PPARα agonists to the nucleus.
By Western blotting, we could detect PPARα in the nucleus but were not able to reveal any PPARα in the cytosol of liver cells. Thus, the nuclear matrix seems to be the predominant compartment for direct L-FABP–PPAR interaction, which, however, could also be possible in the cytosol. Others have shown that PPARα interacts with proteins belonging mainly to the family of transcriptional cofactors (30, 31) via their LXXLL sequence motif (31), which is not found in L-FABP. We surmised that interaction of both proteins could be conferred via the negative charge by ligand bound that protrudes from the binding pocket of L-FABP. Such protruding was shown for oleic acid bound to L-FABP (32). Therefore, we analyzed whether L-FABP–PPARα interaction was affected by ligands. Neither by in vitro pull-down assay under thermodynamic equilibrium conditions nor by mammalian two-hybrid system could we demonstrate a dependence of interaction on ligand binding. A ligand upon binding may, however, create a specific binding motif for protein–protein contacts and thus influence the kinetics of interaction; identification of this motif should be the subject of further studies. It is interesting to speculate whether such protein–protein contacts are mandatory for fatty acid transfer from L-FABP to PPAR in vivo. Once the binding motifs on both proteins are identified, transactivation assays in HepG2 cells with the proteins having mutated binding motifs may give an answer.
Does the fatty acid or drug bound by L-FABP have an effect on the nuclear localization of the binding protein? First, L-FABP with respect to size, with or without ligand, might diffuse freely through the nuclear pores. Although on a protein-based scale the L-FABP concentration in the nucleus is 100-fold below that in the cytosol (11), concentrations related to volume may be different. Second, ligand bound in a manner mentioned above may furnish a recognition signal for targeting the binding protein to the nucleus; third, covalent modification of L-FABP may furnish the targeting signal. Modifications of this protein by cysteinylation and glutathionylation were recognized by us earlier (33). A very recent report by Lawrence et al. (34) strongly favors option two. Those authors show, on the one hand, that L-FABP binds directly to nuclei of rat hepatocytes in a ligand-dependent manner and, on the other hand, that L-FABP, also ligand-dependent, binds to nuclear proteins other than PPARs. A hypothesis for alternative transfer of ligand from L-FABP to PPAR as alluded to above could be that the ligand is released in the latter process to be taken up by PPAR.
The ligand slopes obtained in L-FABP concentration-dependent transactivation experiments (Fig. 4) correlate with the respective relative CAT expressions shown in Fig. 1 and reflect the transactivation power of the respective ligand. Furthermore, extrapolation of the fitted graphs to zero L-FABP concentration leads to a complete loss of PPAR activation. This implies that transactivation is not possible without L-FABP as transporter and strengthens the argument that L-FABP is required for PPARα activation in the hepatocyte-derived HepG2 cells. From the physiological point of view, the concentration of L-FABP in rat liver was reported to be around 70 nmol/g, whereas the concentrations of unesterified fatty acids ranged from 50 to 100 nmol/g (16), suggesting that L-FABP is able to bind most of unesterified fatty acids and to transport them either to their places of metabolic utilization or to PPARα and PPARγ for activation. Recent binding studies performed with a fluorescence displacement assay in our laboratory showed that PPARα is a high-affinity protein for peroxisome proliferators (in the 10 nM range), with lower affinities for fatty acids (in the 100 nM range) (4). L-FABP, in contrast, binds fatty acids with high affinity (10 nM) in the first binding site, with low affinity in the second (100–500 nM) and peroxisome proliferators with affinities around 1 μM (14). Thus, PPARα would be able to displace peroxisome proliferators from both binding sites of L-FABP, whereas fatty acids would preferentially be displaced from the second binding site.
Because PPARγ is found only in low amounts in hepatocytes, the question arises whether the regulation of this receptor by L-FABP is important in this cell type (8). However, because L-FABP is codistributed in kidney, intestine, and brown adipose tissue with PPARγ (8, 35) a possible complex formation of the two proteins in these tissues might be involved in gene regulation.
Taken together, the results reported give compelling evidence that L-FABP in hepatocytes plays a role in fatty acid and drug signaling to affect PPARα activation. As indicated in Fig. 5, L-FABP functions as a mandatory cytosolic gateway for transport of the activators into the nucleus and directly interacts with PPARs. It is tempting to hypothesize that the phenomenon observed for L-FABP and PPARα and PPARγ might be a general principle applicable to other combinations of FABP types and PPAR isoforms. Indeed, a recent report may indicate a stimulation of PPARγ transactivation by adipocyte (A-)FABP (36). It has not escaped our attention that the various FABPs and further structurally related proteins, which are functionally adapted to the specific needs of the cell where they are expressed, could serve as new discriminating targets for nutrients and xenobiotics designed to affect gene transcription.
Acknowledgments
Technical assistance of S. Lütke-Enking in cell culturing is gratefully acknowledged. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 310/A4. C.W. is grateful for a scholarship from the Unilever Stiftungsfonds.
Abbreviations
- ETYA
5,8,11,14-eicosatetraynoic acid
- L-FABP
liver fatty acid binding protein
- PPAR
peroxisome proliferator-activated receptor
- RXR
retinoid X receptor
- CAT
chloramphenicol acetyltransferase
- β-Gal
β-galactosidase
- PPRE
peroxisome proliferator-responsive element
References
- 1.Bocos C, Göttlicher M, Gearing K, Banner C, Enmark E, Teboul M, Crickmore A, Gustafsson J A. J Steroid Biochem Mol Biol. 1995;53:467–473. doi: 10.1016/0960-0760(95)00093-f. [DOI] [PubMed] [Google Scholar]
- 2.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krey G, Braissant O, L'Horset F, Kalkhoven E, Perroud M, Parker M G, Wahli W. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 4.Ellinghaus P, Wolfrum C, Assmann G, Spener F, Seedorf U. J Biol Chem. 1999;274:2766–2772. doi: 10.1074/jbc.274.5.2766. [DOI] [PubMed] [Google Scholar]
- 5.Issemann I, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 6.Wolfrum C, Ellinghaus P, Fobker M, Seedorf U, Assmann G, Börchers T, Spener F. J Lipid Res. 1999;40:708–714. [PubMed] [Google Scholar]
- 7.Wahli W, Devchand P R, Ijpenberg A, Desvergne B. Adv Exp Med Biol. 1999;447:199–209. doi: 10.1007/978-1-4615-4861-4_19. [DOI] [PubMed] [Google Scholar]
- 8.Braissant O, Wahli W. Endocrinology. 1998;139:2748–2774. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- 9.Ockner R K, Manning J A, Poppenhausen R B, Ho W K. Science. 1972;177:56–58. doi: 10.1126/science.177.4043.56. [DOI] [PubMed] [Google Scholar]
- 10.Hohoff C, Spener F. Fett/Lipid. 1998;100:252–263. [Google Scholar]
- 11.Bordewick U, Heese M, Börchers T, Robenek H, Spener F. Biol Chem Hoppe-Seyler. 1989;370:229–238. doi: 10.1515/bchm3.1989.370.1.229. [DOI] [PubMed] [Google Scholar]
- 12.Cannon J R, Eacho P I. Biochem J. 1991;239:781–791. doi: 10.1042/bj2800387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolf B, Oudenampsen-Krüger E, Börchers T, Færgeman N J, Knudsen J, Lezius A, Spener F. Biochim Biophys Acta. 1995;1259:245–253. doi: 10.1016/0005-2760(95)00170-0. [DOI] [PubMed] [Google Scholar]
- 14.Wolfrum C, Börchers T, Sacchettini J C, Spener F. Biochemistry. 2000;39:1469–1474. doi: 10.1021/bi991638u. [DOI] [PubMed] [Google Scholar]
- 15.Kaikaus R M, Chan W K, Ortiz de Montellano P R, Bass N M. Mol Cell Biochem. 1993;123:93–100. doi: 10.1007/BF01076479. [DOI] [PubMed] [Google Scholar]
- 16.Bass N M. Mol Cell Biochem. 1993;123:191–202. doi: 10.1007/BF01076492. [DOI] [PubMed] [Google Scholar]
- 17.Wolfrum C, Buhlmann C, Rolf B, Börchers T, Spener F. Biochim Biophys Acta. 1999;1437:194–201. doi: 10.1016/s1388-1981(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 18.Juge-Aubry C, Pernin A, Favez T, Burger A G, Wahli W, Meier C A, Desvergne B. J Biol Chem. 1997;272:25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- 19.Brandt J M, Djouadi F, Kelly D P. J Biol Chem. 1998;273:23786–23792. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 20.Yu K, Bayona W, Kallen C B, Harding H P, Ravera C P, McMahon G, Brown M, Lazar M A. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 21.Stümpfle K J, Koptides M, Karinch A M, Floros J. BioTechniques. 1996;21:48–50. doi: 10.2144/96211bm09. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee R, Jow L, Noonan D, McDonnell D P. J Steroid Biochem Mol Biol. 1994;51:157–166. doi: 10.1016/0960-0760(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 23.Hubbell T, Behnke W D, Woodford J K, Schroeder F. Biochemistry. 1994;33:3327–3334. doi: 10.1021/bi00177a025. [DOI] [PubMed] [Google Scholar]
- 24.Hertz R, Berman I, Bar-Tana J. Eur J Biochem. 1994;221:611–615. doi: 10.1111/j.1432-1033.1994.tb18773.x. [DOI] [PubMed] [Google Scholar]
- 25.Tugwood J D, Issemann I, Anderson R G, Bundell K R, McPheat W L, Green S. EMBO J. 1992;11:433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardot O, Aldridge T C, Latruffe N, Green S. Biochem Biophys Res Commun. 1993;192:37–45. doi: 10.1006/bbrc.1993.1378. [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto T, Fujita T, Usuda N, Cook W, Qi C, Peters J M, Gonzalez F J, Yeldandi A V, Rao M S, Reddy J K. J Biol Chem. 1999;274:19228–19236. doi: 10.1074/jbc.274.27.19228. [DOI] [PubMed] [Google Scholar]
- 28.Lefèbvre A M, Peinado-Onsurbe J, Leitersdorf I, Briggs M R, Paterniti J R, Fruchart J C, Fievet C, Auwerx J, Staels B. Arterioscler Thromb Vasc Biol. 1997;17:1756–1764. doi: 10.1161/01.atv.17.9.1756. [DOI] [PubMed] [Google Scholar]
- 29.Vu-Dac N, Chopin-Delannoy S, Gervois P, Bonnelye E, Martin G, Fruchart J C, Laudet V, Staels B J. J Biol Chem. 1998;273:25713–25720. doi: 10.1074/jbc.273.40.25713. [DOI] [PubMed] [Google Scholar]
- 30.Mizukami J, Taniguchi T. Biochem Biophys Res Commun. 1997;240:61–64. doi: 10.1006/bbrc.1997.7602. [DOI] [PubMed] [Google Scholar]
- 31.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J, Winter N, Terwey D, Bratt J, Banaszak L. J Biol Chem. 1997;272:7140–7150. doi: 10.1074/jbc.272.11.7140. [DOI] [PubMed] [Google Scholar]
- 33.Dörmann P, Börchers T, Korf U, Højrup P, Roepstorff P, Spener F. J Biol Chem. 1993;268:16286–16292. [PubMed] [Google Scholar]
- 34.Lawrence J W, Kroll D J, Eacho P I. J Lipid Res. 2000;41:1390–1401. [PubMed] [Google Scholar]
- 35.Glatz J F, Börchers T, Spener F, van der Vusse G J. Prostaglandins Leukotrienes Essent Fatty Acids. 1995;52:121–127. doi: 10.1016/0952-3278(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 36.Hertzel A V, Bernlohr D A. Mol Cell Biochem. 1998;188:33–39. [PubMed] [Google Scholar]