Abstract
AgBase (http://www.agbase.msstate.edu/) provides resources to facilitate modeling of functional genomics data and structural and functional annotation of agriculturally important animal, plant, microbe and parasite genomes. The website is redesigned to improve accessibility and ease of use, including improved search capabilities. Expanded capabilities include new dedicated pages for horse, cat, dog, cotton, rice and soybean. We currently provide 590 240 Gene Ontology (GO) annotations to 105 454 gene products in 64 different species, including GO annotations linked to transcripts represented on agricultural microarrays. For many of these arrays, this provides the only functional annotation available. GO annotations are available for download and we provide comprehensive, species-specific GO annotation files for 18 different organisms. The tools available at AgBase have been expanded and several existing tools improved based upon user feedback. One of seven new tools available at AgBase, GOModeler, supports hypothesis testing from functional genomics data. We host several associated databases and provide genome browsers for three agricultural pathogens. Moreover, we provide comprehensive training resources (including worked examples and tutorials) via links to Educational Resources at the AgBase website.
INTRODUCTION
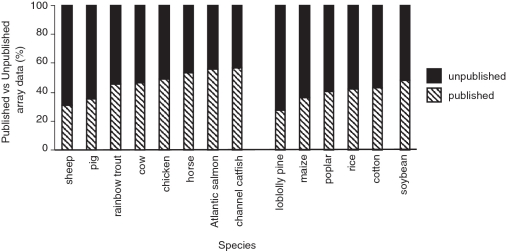
AgBase was founded as several agriculturally important genomes were sequenced or scheduled for sequencing (1). While our initial goal to provide functional modeling resources for agricultural researchers has not changed, advances in ‘omics’ technologies are dramatically changing the way biologists do research, and agriculture is not exempt from this paradigm shift. Data acquisition is no longer an impediment for ‘omics’ experiments; instead the focus is shifting to deriving value (i.e. knowledge) from this data (2). For example, there are currently (9 September 2010) 1509 microarray data sets for common agricultural species in the Gene Expression Omnibus (GEO) database (3,4) but only 57% are published and the proportion of published data varies widely between species (Figure 1). This is exacerbated by data sets that have not yet been submitted to public databases, the development of new arrays for agricultural species [e.g. horse (5) and turkey (6)] and the advent of RNA-Seq. Researchers who wish to model their functional genomics data sets are becoming more reliant on resources that provide annotated data.
Figure 1.
Publication of agricultural microarray data. On average, less than half of the agricultural microarray data sets submitted to the NCBI GEO database are published. These statistics are based upon gene expression data submitted to the NCBI GEO database. Records are shown for as at 9 September 2010. For each species the number of expression data sets linked to a PubMed record was used to determine the proportion of unpublished data sets. Note that this approach is likely to under-estimate the amount of unpublished data sets as it does not include data not submitted to this database.
While each agricultural species has its own published information that can be utilized for functional modeling, analysis of data from literature is not easily done at an ‘omics’ scale. To overcome this limitation, annotation is used to link biological knowledge to biological data. While manual biocuration of literature provides detailed, organism specific, high quality annotation, this process is necessarily slow and current funding cannot enable manual curation to keep pace with the increasing rate of data acquisition. Moreover, many sequences have no associated literature and can only be annotated based upon computational sequence analysis [e.g. novel transcriptional elements identified by RNA-Seq (7)]. Instead, biocurator time needs to be used efficiently and target high impact data in the literature. Moreover, biocurators provide necessary checks for computational annotation [e.g. mapping files used by computational pipelines (8) and rules for applying annotations across species]. AgBase uses a mixture of manual and computational biocuration to provide the necessary annotation to support research.
The following sections focus on new developments to AgBase in response to changes in the way researchers are applying functional genomics to agriculturally important species. We will highlight developments in the type of data available via AgBase, changes to the interface and tools, education and training initiatives and future directions.
AgBase DATABASE
AgBase is implemented using MySQL as the relational database management system running on a Linux server, using an Apache web server, and Perl CGI scripts as the web interface. The AgBase database combines data from the AgBase biocurators along with external data from UniProt, the Gene Ontology (GO) Consortium, NCBI and Affymetrix (Supplementary Data S1). The AgBase database is updated every two months.
THE AgBase WEB INTERFACE
The AgBase web interface is redesigned using graphic design principles to enhance the user’s ability to navigate the site. To direct users based upon function and species the new interface features a drop down menu across the top of the page featuring the most commonly accessed pages. The left sidebar menu has links to additional resources including download files and educational resources. Additional data sources that we host are also featured on the right side of the AgBase homepage.
The AgBase homepage allows researchers to search the content of the database using either public database accessions/identifiers (from UniProtKB, Genbank and GO) or protein or gene names/symbols. Users may choose to select all of AgBase or limit their search to one of the 18 species that AgBase is actively supporting. In addition to these species, AgBase includes external GO annotations provided by other GO Consortium members. This enables users to search, for example, mouse or yeast records in addition to the agricultural species linked in AgBase. Agriculturally relevant species also have their own dedicated web pages that can be accessed from the menu at the top of the page. Species pages include links to organism specific community resources, the gene association file (GAF) provided by AgBase and GO annotation statistics (via the GOProfiler link) for the species as well as species specific text and BLAST search links. There are specific cases where the species page incorporates more than one taxon; e.g. cotton and rice, as GO annotations are distributed across several closely related taxonomies. Species pages include taxonomy information used to gather the information for the page. Researchers are encouraged to contact AgBase to request the addition of a species page.
Database queries are now the more flexible. We added the ability to search using protein Genbank identifiers (i.e. ‘gi numbers’) because proteomics data sets may be reported as gi numbers and because several agricultural species are not well represented using UniProtKB accessions/identifiers. Alternatively, users may select an unspecified ID search to search for all supported identifier types. The Gene Name search now includes (or excludes) either synonym matches or wildcard matches. Since very few agriculturally important species have standardized gene nomenclature projects, this expanded search capacity helps researchers identify their genes/proteins of interest. AgBase biocurators make every effort to clarify gene nomenclature where possible but without recognized gene nomenclature authorities this information is not easily disseminated.
Guidance for using the AgBase tools includes help notes and a series of worked tutorials that we update with each training workshop. As with any online resource, we rely on user’s input for continual improvements to our help notes. We encourage users to contact AgBase directly for either assistance or comment that we may continue to improve our ability to assist researchers. We also encourage researchers to add their own data or request annotations based upon publications they know to be linked to their gene(s) of interest. A Community Request/Submission page allows users to either Request or Submit GO annotations to AgBase. Sequences that are not yet in public databases may be GO annotated by contacting AgBase directly and will be held from public release until notification. GO annotations submitted by researchers are checked by biocurators and then quality checked prior to release in AgBase. The researcher who submitted the GO annotation is credited for the GO annotation using the standard GAF field ‘Assigned_by’.
DATA TYPES, SOURCE AND ANNOTATION STRATEGIES
AgBase biocurators currently provide 590 240 GO annotations for 105 454 gene products from 64 species (as of 10 August 2010). These AgBase derived annotations are made available as two different GAFs, which are both quality checked prior to release. The GO Consortium (AgBase GOC) GAF contains annotations released to the GO Consortium (9). A second GAF (AgBase Community) contains:
annotations for gene products not supported by the European Bioinformatics Institute GOA (EBI GOA) Project (e.g. transcripts and Genbank ‘predicted’ proteins);
‘Inferred from Sequence Similarity’ (ISS) annotations to evidence codes no longer accepted as of June 2007 (note that these annotations are updated during standard QC procedures); and
annotations from community researchers, where the source of the annotation is attributed in each case.
Note that the AgBase Community GAF contains GO annotations that have not yet been submitted to the GO Consortium. However, both AgBase GAFs are fully compliant with the 17-column GAF format (GAF2.0) implemented by the GO Consortium (1 June 2010). AgBase also provides species specific GAFs for 18 agricultural organisms, which are a comprehensive source of GO annotations derived from both AgBase and other GO Consortium members. Since we are currently funded to provide literature based GO annotations for chicken, bovine, maize and cotton, the gene products we annotate are predominantly from these species. However, our GO annotations also include other gene products from agriculturally important species where GO annotation was requested by AgBase users (e.g. pig, horse, dog) and incidental GO annotations for other species’ gene products described in literature that we biocurated for chicken, bovine, maize and cotton. We are also biocurating plant gene products using the Plant Ontology (10).
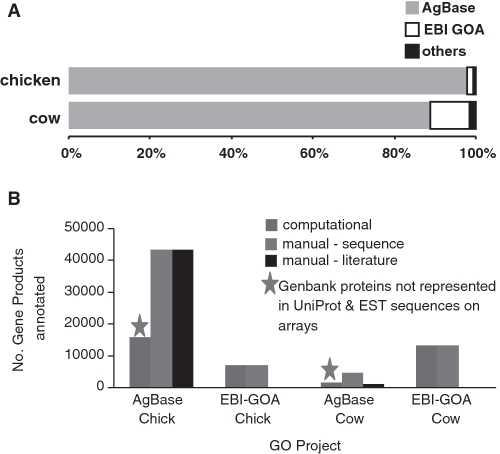
The annotations provided by AgBase are either computationally derived or manually curated from literature. This dual annotation strategy enables us to capture the ‘breadth’ of GO annotation for agricultural gene products (by computational methods) as well as the ‘depth’, or detailed organism specific functional information (via literature curation). We use InterProScan (11) to provide IEA (‘inferred from electronic annotation’) annotations for agricultural ESTs and ‘predicted’ gene products based on functional motifs and domains. Since both AgBase and EBI GOA provide GO annotations for chicken and cow gene products, our aim is to provide complementary GO annotations for these two species. While EBI GOA provides IEA annotations for proteins in UniProt, we provide IEA annotations for proteins not represented in UniProt and transcripts represented on commonly used arrays (Figure 2). We provide additional annotation by identifying strict 1:1 orthologous genes and transferring GO from the better annotated gene (typically from a model organism e.g. human or mouse) to its orthologous gene product. When this method of GO annotation is manually reviewed by biocurators it is assigned the ISO (inferred from sequence ortholog) evidence code; GO annotations that are automatically transferred are assigned an IEA evidence code, as mandated by GO Consortium evidence code guidelines. GO identifiers that are computationally transferred to a gene product in another species are manually reviewed during the QC process to ensure that the transfer is biologically appropriate.
Figure 2.
GO annotation strategies for AgBase and the EBI’s Gene Ontology Annotation (EBI GOA) group. Both AgBase and the EBI GOA Project provide GO annotations for chicken and cow. This annotation effort is complementary. (A) The percentage of biocuration for each group is shown for literature biocuration projects for chicken and cow. (B) Complementary biocuration of chicken and cow gene products. The EBI GOA Project provide computational GO annotation for UniProtKB records while the AgBase computational annotation effort focuses on Genbank gene products not represented in the UniProtKB database (typically these are gene products represented on commonly used arrays).
Since manual biocuration of the literature to provide GO annotation is necessarily slower, we target our annotation based upon user requests and gene products represented on commonly used microarrays. We provide ID mapping and GO annotation files for commonly used chicken and bovine arrays (Supplementary Data S2). AgBase biocurators target manual biocuration using a Gene Prioritization interface that ranks genes based upon user requests or presence on microarrays. When researchers request annotations via the AgBase Community Requests & Submissions page, they are able to access the Gene Prioritization list to determine where their request is in the queue. (Note that when we biocurate a paper we provide GO annotations for all gene products represented in that paper, regardless of species; if another GO Consortium group is already providing GO annotation for a species this information is forwarded to that group.) Another novel tool that we use to focus our manual biocuration effort is the extracting Genic Information From Text tool (eGIFT) (12). eGIFT searches PubMed to identify literature containing functional information and suggests GO terms that are likely to be present in these publications. Integrating eGIFT with our biocuration interface enables AgBase biocurators to rapidly identify publications for GO annotation. Since details of papers we have biocurated are also made available via the Journal Database (JDB) (1), we also integrated the JDB with our biocuration interface. Since we use the JDB to record publications that we could not access or that were biocurated but contained no GO annotation, this information can be now be captured directly from the biocuration interface and viewed in the JDB. When the reviewed publication does not contain GO annotation, biocurators submit functional information to the National Center for Biotechnology Information (NCBI) Gene Reference Into Function (GeneRIF;). This allows us to capture additional information (e.g. tissue expression, protein structure, post-translation modifications and structural annotation); chicken, cow and maize species are well represented amongst the GeneRIFs entries (with chicken and bovine in the top 12 and maize ranked number 30 of 984 species with GeneRIF records). We encourage researchers to make use of the NCBI GeneRIF interface to ensure that their publications are linked to the appropriate gene(s).
In addition to providing GO annotations for the agricultural research community, we also provide structural annotations and host other genome related databases. The structural annotations at AgBase are reached via the Proteogenomics page and, instead of the more traditional gene model annotations, are provided as proteogenomic mapping results for chicken and several microbial species using. Proteogenomic mapping is a method for using proteomics data for improved genome annotation (13,14). Using this method, mass spectra data is searched against the genome translated in all six reading frames and matches that do not coincide with known genes are used to generate Expressed Protein Sequence Tags (ePSTs) (15). These ePSTs represent translated regions of the genome, many of which are novel. More information about proteogenomic mapping, ePSTs and how these resources can be used to improve structural annotation of the genome is provided (Supplementary Data S3). Briefly, a GMOD genome browser (16) provides visualization of ePSTs for the microbial species and we are in the process of providing a genome browser to support visualization of eukaryotic ePSTs. We will use the eukaryotic based genome browser to visualize chicken ePSTs and RNA-Seq tags that we are currently identifying from multiple chicken tissues.
TOOLS TO SUPPORT FUNCTIONAL MODELING OF AGRICULTURAL RESOURCES
We recently published a quantitative experiment demonstrating the essentiality of up-to-date functional annotation for modeling functional genomics data sets; failure to update functional annotation results in inaccuracies in ‘omics’ data modeling (17). A key role of AgBase is to provide GO annotations for agricultural gene products and facilitate GO-based modeling in agriculturally important species. While there are many tools and resources available for functional modeling, few support agricultural species. Our approach to tool development is two-fold: (i) provide the data to support existing functional modeling tools and (ii) develop additional tools to bridge gaps between existing modeling tools. The AgBase Tools Overview page groups tools based on functional categories: Functional Analysis Using GO, Array Analysis, Proteomics Analysis and Sequence Analysis.
Providing data to support functional modeling
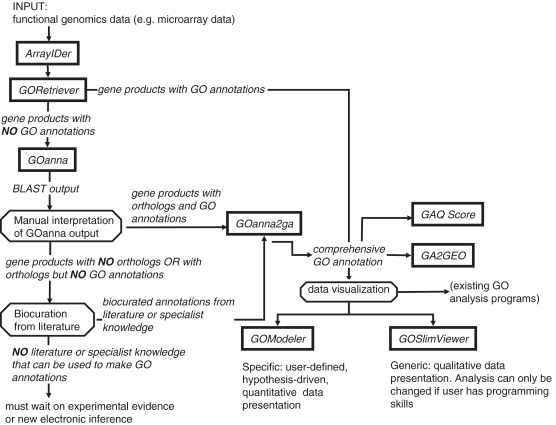
Most tools grouped in the category ‘Functional Analysis Using GO’ may be used independently, or as a pipeline (Figure 3) to provide GO annotations for experimental data sets. The use of these tools as a pipeline to rapidly add GO to a data set enables researchers to do functional modeling when there is little or no GO annotation available for their data set. One of these tools, GOanna, was developed when there were very few tools that would use BLAST searches to add GO to homologous sequences, and was the only tool that allowed users to scan the BLAST alignments to determine good matches (1). While there are now several other tools that use the same approach (18,19), this tool remains one of the most highly accessed tools at AgBase. GOanna now utilizes an updated version of BLAST, more accession types and customized databases (Supplementary Data S4). A complementary tool, GOanna2ga, converts the GOanna output file to standard GAF format and a truncated GOSummary file format that is supported by GOSlimViewer. The GAF can be used in GO enrichment analysis tools that allow users to upload additional GO annotations [e.g. BiNGO (20), GOStat (21), Onto-Express (22,23)].
Figure 3.
AgBase tools for functional analysis using the GO. This figure shows the individual AgBase tools for functional analysis of the GO and how they can be used sequentially as a pipeline to provide GO annotations and functional modeling for data sets. Square boxes represent AgBase tools; octagons represent manual steps or checks in the process.
Since microarrays are commonly used in agricultural based functional genomics, we provide tools to assist with microarray analysis. The Array GO Mapper Tool (AGOM) (24) leverages annotations data associated with Affymetrix GeneChip arrays. Users can input a list of accessions and AGOM checks these accessions against the ID mapping data provided with the Affymetrix array and returns available annotation, including GO annotation. This enables researchers using Affymetrix arrays to rapidly access annotation for their data and, more importantly, users who have a less well annotated array to rapidly retrieve ID mappings and annotation to begin their functional modeling.
AgBase also supports more general tools for sequence analysis. The MSVIS tool provides a new approach for simultaneous visualization of conserved motifs and sequence alignment (25). A genome wide approach to sequence analysis is the Proteogenomic Mapping Pipeline, which uses high-throughput liquid chromatography mass spectrometry proteomics to complement computational structural genome annotation (1,26). This tool is now modified to enable its use for annotating larger eukaryotic genomes.
Bridging tools for functional modeling
Since many agricultural genomes have poor annotation compared to model organisms, we provide tools to help agricultural researcher’s access existing resources and tools for modeling their data. The GOProfiler tool enables researchers quantify the amount of GO annotation that is available for their species of interest (1). GOProfiler counts GO annotations based upon taxon ID for all GO annotations submitted to the GO Consortium and the AgBase Community file. Researchers enter the taxon ID or use the Taxonomy Browser to find the taxon ID for their species. Both the number of GO annotations and the number of gene products with GO annotations are reported, with GO annotations also displayed based upon GO Evidence Codes. We also report the number of unannotated gene products based upon protein entries in the UniProtKB database. A direct link to the relevant GOProfiler summary table is available from each of the AgBase organism pages. While GOProfiler provides an overview of GO annotation available for entire species, the GO Annotation Quality Score (GAQ Score) provides a quantitative assessment of GO annotation for a particular data set (27). We provide GAQ Scores for each array we have annotated to help researchers assess the functional annotation available for these arrays, enabling researchers to include a consideration of functional modeling in their array selection process at the beginning of their experiment. Moreover, researchers can use the online GAQ Score tool to calculate GAQ Scores for their own data sets by entering a GAF (GAF 2.0 format). This provides a rapid way to assess the impact of adding your own GO annotations to an experimental data set using, for example GOanna.
The most common support request received at AgBase is for assistance mapping public database IDs so that data sets can be changed to an ID type supported by functional modeling tools. This is hardly surprising given the proliferation of biological databases (2,28) and several databases and resources already provide tools for mapping between public database accessions. Notable amongst these for their ease of use, accessibility and ability to map a broad range of database IDs and are the Ensembl BioMart data mining (29), UniProt ID mapping (30) and DAVID Gene ID Conversion tools (31). To supplement these tools we provide ArrayIDer (32), a tool that has the advantage of including NCBI dbEST accessions. Although many agricultural arrays are based upon EST sequences, few (if any) functional modeling tools support EST accessions, creating a gap for researchers wishing to model data sets produced using these arrays. ArrayIDer now accepts multiple ID types including EST accessions and returns a table of the input accessions and equivalent mappings to genes, transcripts and proteins from NCBI/EMBL/DDBJ, Ensembl and UniProt. We also provide AffyID, a tool for ID mapping based on Affymetrix Probe set IDs. While there are several existing tools that map Affymetrix Probe set IDs to public database IDs, it is important to note that for agricultural based arrays in particular, Affymetrix annotation files are not updated as frequently as model organism arrays (Table 1). Since AgBase biocurators are providing updated ID mappings and GO annotations for agricultural arrays, AffyID uses this updated data.
Table 1.
Annotation updates for agricultural arrays
| Platform ID | Array name | Submitted | Last update |
|---|---|---|---|
| Chicken | |||
| GPL3213 | Affymetrix Chicken Genome Array | November 2005 | June 2009 |
| GPL5480 | ARK-Genomics G. gallus 20K v1.0 | July 2007 | July 2007 |
| GPL1731 | DEL-MAR 14K Integrated Systems | December 2004 | March 2006 |
| Bovine | |||
| GPL2853 | UIUC Bos taurus 13.2K 70-mer oligoarray | September 2005 | March 2007 |
| GPL2864 | UIUC Cattle 7,872-element cDNA - alternate version | September 2005 | March 2007 |
| GPL2112 | Affymetrix Bovine Genome Array | May 2005 | June 2009 |
| Pig | |||
| GPL7435 | Swine Protein-Annotated Oligonucleotide Microarray | October 2008 | November 2008 |
| GPL3608 | DIAS_PIG_55K3_v1 | March 2006 | May 2009 |
| GPL1881 | Qiagen-NRSP-8 porcine oligo array | February 2005 | May 2005 |
| Horse | |||
| GPL10248 | Agilent 4x44k Horse Gene Expression microarrays | March 2010 | March 2010 |
| GPL8582 | MacLeod custom equine cartilage 10K cDNA microarray version 3 | May 2009 | October 2009 |
| Maize | |||
| GPL4032 | Affymetrix Maize Genome Array | July 2006 | June 2009 |
| GPL3538 | SAM3.0 | March 2006 | November 2006 |
| GPL3333 | SAM1.1a | January 2006 | March 2006 |
| GPL1996 | Maize cDNA Generation II Version B | April 2005 | May 2005 |
| Rice | |||
| GPL1829 | Rice Genome Oligo Set V1.0 | January 2005 | October 2008 |
| GPL892 | Agilent-012106 Rice Oligo Microarray G4138A | January 2004 | September 2008 |
| GPL8161 | NSF Rice Oligonucleotide Array 45K One Chip Version | February 2009 | February 2009 |
| Soybean | |||
| GPL3015 | Keck Glycine max 18kA cDNA Prints101-108 | October 2005 | October 2005 |
| GPL1012 | Gm-r1088 | February 2004 | May 2005 |
| GPL229 | Gm-r1070 | December 2002 | October 2005 |
| Tomato | |||
| GPL9923 | CombiMatrix 90K TomatArray 1.0 | January 2010 | August 2010 |
| GPL4741 | Affymetrix Tomato Genome Array | January 2007 | June 2009 |
| GPL3034 | Cornell-CGEP Tomato 13K vTOM1 | October 2005 | November 2005 |
Arrays for agricultural species with the greatest numbers of data sets submitted to the NCBI GEO database (as at 9 September 2010) are shown, along with information about when the array platform data was submitted and its last update. Updates typically include ID mapping; updated functional information for transcripts represented on arrays is not always included and is harder to assess collectively.
A common starting point for functional modeling is to use GO Slim sets to provide a high level summary of GO function for a particular data set (i.e. a highly summarized view of the associated GO using extremely broad functional terms). For example, the GO currently contains 32,284 GO terms (ontology version 1.1394, 27/08/10) but the PIR GOSlim contains only 467 of these while the GOA GOSlim contains 62. GOSlimViewer enables researchers to use these GOSlim sets to summarize the GO annotation for their data (26). Based upon user requests, we modified this tool to include additional GOSlim sets and to provide detailed information about how individual gene products and their GO annotations are summarized. GOSlimViewer now supports the PIR GOSlim set and the Biological Process slim set developed specifically for prokaryotes by researchers at The Institute for Genomic Research (TIGR), now the J. Craig Venter Institute (JCVI). In addition to the summarized function for each ontology, GOSlimViewer results now also include a link to ‘View accessions for each slim id’. This link shows each summarized GO:ID for the data set, the gene products summarized to this GO term and their original annotation GO:ID. This enables the user to identify the entries that contributed to the summarized functional groups.
Summarizing data based upon GOSlim sets differs from GO enrichment analysis tools as it does not determine whether or not particular GO terms are over/under-represented in the experimental data set. Very many GO enrichment analysis tools exist and several are expanding their capacity to support new species (including agricultural species) or are specifically designed to support functional modeling of agricultural data (33). Our novel approach to using the GO for functional modeling is GOModeler, which enables hypothesis testing of gene expression data (34). GOModeler enables the researcher to ‘translate’ hypothesis statements (or expected phenotypes) into equivalent GO terms which are then scored for their effect on each gene in an expression data set (pro, anti, no effect). The user’s gene expression data is overlaid onto this scoring matrix and summed for each hypothesis statement to determine overall effects for each hypothesis statement. This tool relies on researcher’s having expert biological knowledge and it does not do a ‘black-box’ or undirected GO enrichment analysis (like many researchers commonly use); therefore, we provide both detailed online help and an online tutorial for GOModeler.
AgBase currently provides two tools to support high throughput proteomics research. PepFly allows researchers to predict proteolytic peptides from tandem mass spectrometry samples that are likely to be observed (35). This tool enables researchers to calculate protein coverage based upon experimental conditions. ProtQuant allows protein quantification from isotope label-free proteomics data sets (36).
USING AgBase FOR ‘OMICS’ DATA SET FUNCTIONAL MODELING
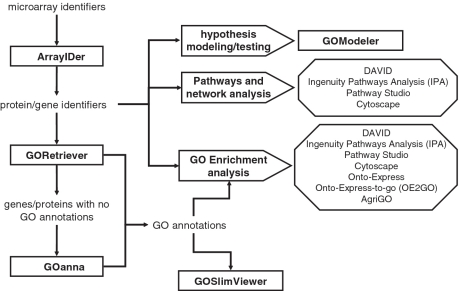
While users can search the AgBase website using individual gene products, species, sequences or the GO, the website is specifically designed for analyzing functional genomics data. Our paradigm is that modeling is driven by the biological system, technological platform used to derive the experimental data and, most importantly, by the expert experimentalist. Typically, functional modeling approaches include (i) grouping by function (e.g. using GOSlim sets); (ii) functional enrichment analysis (including GO enrichment); (iii) pathway and network analysis and (iv) hypothesis testing (Figure 4). Available functional modeling tools may combine these different approaches, for example many tools combine (ii) and (iii) and the data obtained from these different approaches is often complementary. As previously mentioned, functional analysis often requires researchers to map their data to a public database accession accepted by these tools and in species where there is little or no GO available, add GO to support functional modeling. Adding additional annotation can considerably change the outcome of functional modeling (17).
Figure 4.
Overview of functional modeling strategies. This figure shows how the AgBase tools can be used as part of a larger functional modeling strategy that incorporates other, existing functional analysis tools. Square boxes represent AgBase tools; arrow shaped boxes represent overall modeling approaches and octagonal boxes contain representative example of non-AgBase tools that are commonly used for functional modeling.
RESOURCES HOSTED BY AgBase
AgBase hosts several agricultural based databases. The Bovine Gene Expression Atlas (BGA) is a rapidly expanding compendium of over 7 million expressed sequences from 81 different bovine tissues (37). These sequence tags are visualized using GBrowse bovine genome build 3.1. The BGA, which allows researchers to search for landmarks (e.g. genes) or regions of the genome to identify expression patterns and to specify tissue expression, facilitates analysis of gene expression data and enables researchers to link gene expression to gene function.
The Corn Fungal Resistance Associated Sequences (CFRAS) database integrates data from expression, genetic mapping and sequencing, enabling researchers to simultaneously examine many lines of evidence and evaluate the potential role of a gene or a group of maize genes when exposed to Aspergillus flavus infection and aflatoxin production (38). This facilitates the identification of molecular markers for producing corn hybrids with increased resistance to aflatoxin accumulation.
AgBase also hosts the Chicken Gene Nomenclature Committee (CGNC) database. The CGNC is an international group of researchers interested in providing standardized gene nomenclature for chicken genes (39). A Chicken Gene Annotation Tool is already available (http://edit-genenames.roslin.ac.uk/) which assigns chicken nomenclature based on predicted orthology to human genes. The CGNC database hosted by AgBase includes this information and adds manually biocurated nomenclature using AgBase funded chicken biocurators and interested contributors. Both resources are part of a united CGNC effort and nomenclature data is shared and coordinated between these two resources. We strongly encourage researchers with domain knowledge to participate in this nomenclature effort.
The Host-Pathogen Interaction Database (HPIDB) is a unified resource for host-pathogen interactions which integrates experimental protein–protein interactions (PPIs) from several public databases (40). The database can be searched using sequence identifiers, symbol, taxonomy, publication, author, interaction type or using sequences. The taxonomic categorization of proteins (bacterial, viral, fungi, etc.) involved in PPI enables the user to do phyla specific BLASTP searches. In addition, HPIDB allows searching for homologous host-pathogen interactions based on user provided host and/or pathogen proteins.
COMMUNITY OUTREACH AND TRAINING
AgBase personnel are committed to providing ongoing support for the agricultural research community. We do this by providing online Educational Resources, conducting Functional Modeling training workshops, answering user questions directed to the AgBase website and by direct collaboration with agricultural researchers. The Educational Resources provided on the AgBase website include links to general information about the GO and AgBase, presentations about functional modeling and links to our Functional Modeling Workshops.
Functional Modeling Workshops are held by request and are typically hosted by an on-site researcher who serves as the local coordinator. (To request a training workshop, please contact AgBase.) Workshops are tailored to meet the participants’ specific needs (e.g. duration and topics covered) and attendees are encouraged to bring their own data to work on. We also contact and encourage GO tool developers to participate in these workshops by providing tutorials. Via the Educational Resources link we provide a continuous link to materials and resources covered during workshops, including comprehensive access to all presentations, tutorials and worked examples, additional resources requested by participants and links to websites and publications. Users should note that workshop pages are customized for each workshop and not updated afterwards; for self-training purposes we recommend using one of the more recent workshops.
In addition to providing training opportunities and ongoing online support, we also interact with the agricultural research community via direct research collaborations. We worked directly with microarray users and developers to provide ID mapping and GO annotations for the FHCRC chicken 13K (GPL2863) and Equine Whole Genome Oligonucleotide microarrays (5) and are currently working to provide the same data for the 15K Agilent Sheep Gene Expression microarray (019921). We also are working with investigators, post-doctoral associates and students from several institutions to provide genome mapping and/or GO annotation for their RNA-Seq data and improve structural annotation and linkage mapping for the sheep genome. We can and do assist the agricultural research community by using our computational pipelines to provide GO annotation for experimental data sets (including RNA-Seq data), developing new bioinformatics tools, doing direct functional modeling of high-throughput data and doing bioinformatics analyses to support omics strategies.
FUTURE DIRECTIONS
We are continuing to build collaborative links with other biological databases and resource providers to expand AgBase capabilities and integrate our data with existing public resources. We work closely with other member groups of the GO Consortium, particularly the EBI GOA Project (8) and the Reference Genome Project (41) members. AgBase personnel doing chicken biocuration work closely with other BirdBase members (including Gallus GBrowse, GEISHA, AvesWiki), CGNC and NCBI to provide GO annotations and standardized gene nomenclature. We will also begin providing functional annotation for chicken miRNAs and their targets. As we expand our biocuration efforts to agricultural plants we are actively developing collaborative links with Gramene and MaizeGDB to support continued/expanded biocuration of cereal crops. We are aware of the need to utilize high performance computing (HPC) resources and are already using HPC to provide computational based GO annotations and to assist with collaborative projects with agricultural researchers whose research requires bioinformatics support. We also believe that public and private ‘cloud’ computing can be valuable and economic to the research communities and are beginning to build specific HPC capacity.
CONTACTING AgBase
Interaction with the user community is vital for the success of AgBase. We encourage the submission of new data, the correction of errors and ideas for making this database of even greater use to the community (including ideas for new computational tools). AgBase curators make every effort to maintain data integrity by linking data with researchers, references and methods whenever possible. Questions about AgBase, data updates or errors can be addressed to agbase@cse.msstate.edu.
DATABASE AVAILABILITY
AgBase is freely available via the AgBase website. All data is publicly available via this website and is disseminated to public databases as appropriate. Bioinformatic tools at AgBase are either freely available online or, if they are not amenable to online analysis, available for download at the AgBase Tools page.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Mississippi State University—Office of Research (to AgBase); Division of Agriculture and Forestry, College of Veterinary Medicine; Bagley College of Engineering; Life Sciences and Biotechnology Institute and Mississippi Agriculture and Forestry Experiment Station; National Science Foundation project (EPS-0903787 to S.M.B., partial); National Research Initiative of the US Department of Agriculture Cooperative State Research, Education and Extension Service (grant number MISV-329140); National Institutes of Health National Institute of General Medical Sciences (NIGMS) (project 07111084); US Department of Agriculture, Agricultural Research Service (cooperative agreement number 6402-21000-033-01S); US Department of Agriculture National Institute of Food and Agriculture (grant numbers MIS-069270 and MIS-241080). Approved for publication as Journal Article No J11926 of the Mississippi Agricultural and Forestry Experiment Station, Mississippi State University. Funding for open access charge: US Department of Agriculture Cooperative State Research, Education and Extension Service (grant number MISV-329140, in part).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Michelle Gwin Giglio (University of Maryland) for supplying us with the Institute for Genomic Research (TIGR) Prokaryote GOSlim and members of the GO Consortium and GO Reference Genome Project for their continued support. We also acknowledge the work done by members of the Plant Ontology to develop this ontology and assist others with its use. We are grateful to Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) staff for technical assistance with developing gene nomenclature resources for chicken; Janet Weber [National Library of Medicine (NLM)/National Institutes of Health (NIH)/National Center for Biotechnology Information (NCBI)] for allowing us access to National Center for Biotechnology Information (NCBI) annotation resources and her continued help and support; tool developers at Onto-Tools and AgriGO (in particular Purvesh Khatri and Zhen Su) for supporting our training workshops and collaborators Carl Schmidt, Vijay Shanker and Oana Tudor (University of Delaware) for developing eGIFT.
REFERENCES
- 1.McCarthy FM, Bridges SM, Wang N, Magee GB, Williams WP, Luthe DS, Burgess SC. AgBase: a unified resource for functional analysis in agriculture. Nucleic Acids Res. 2007;35:D599–D603. doi: 10.1093/nar/gkl936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, et al. Big data: the future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bright LA, Burgess SC, Chowdhary B, Swiderski CE, McCarthy FM. Structural and functional-annotation of an equine whole genome oligoarray. BMC Bioinformatics. 2009;10(Suppl. 11):S8. doi: 10.1186/1471-2105-10-S11-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lederman L. Microarrays. BioTechniques. 2009;47:659–661. [Google Scholar]
- 7.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 8.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal P, Avraham S, Ilic K, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, et al. Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp. Funct. Genomics. 2005;6:388–397. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tudor CO, Schmidt CJ, Vijay-Shanker K. eGIFT: mining gene information from the literature. BMC Bioinformatics. 2010;11:418. doi: 10.1186/1471-2105-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffe JD, Berg HC, Church GM. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- 14.Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy FM, Cooksey AM, Wang N, Bridges SM, Pharr GT, Burgess SC. Modeling a whole organ using proteomics: the avian bursa of Fabricius. Proteomics. 2006;6:2759–2771. doi: 10.1002/pmic.200500648. [DOI] [PubMed] [Google Scholar]
- 16.Nanduri B, Wang N, Lawrence ML, Bridges SM, Burgess SC. Gene model detection using mass spectrometry. Methods Mol. Biol. 2010;604:137–144. doi: 10.1007/978-1-60761-444-9_10. [DOI] [PubMed] [Google Scholar]
- 17.van den Berg BH, McCarthy FM, Lamont SJ, Burgess SC. Re-annotation is an essential step in systems biology modeling of functional genomics data. PLoS One. 2010;5:e10642. doi: 10.1371/journal.pone.0010642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 19.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 21.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 22.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, Tarca AL, Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buza TJ, Kumar R, Gresham CR, Burgess SC, McCarthy FM. Facilitating functional annotation of chicken microarray data. BMC Bioinformatics. 2009;10(Suppl. 11):S2. doi: 10.1186/1471-2105-10-S11-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankun-Kelly TJ, Lindeman AD, Bridges SM. Exploratory visual analysis of conserved domains on multiple sequence alignments. BMC Bioinformatics. 2009;10(Suppl. 11):S7. doi: 10.1186/1471-2105-10-S11-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy FM, Wang N, Magee GB, Nanduri B, Lawrence ML, Camon EB, Barrell DG, Hill DP, Dolan ME, Williams WP, et al. AgBase: a functional genomics resource for agriculture. BMC Genomics. 2006;7:229. doi: 10.1186/1471-2164-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buza TJ, McCarthy FM, Wang N, Bridges SM, Burgess SC. Gene Ontology annotation quality analysis in model eukaryotes. Nucleic Acids Res. 2008;36:e12. doi: 10.1093/nar/gkm1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cochrane GR, Galperin MY. The 2010 Nucleic Acids Research Database Issue and online Database Collection: a community of data resources. Nucleic Acids Res. 2010;38:D1–D4. doi: 10.1093/nar/gkp1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.van den Berg BH, Konieczka JH, McCarthy FM, Burgess SC. ArrayIDer: automated structural re-annotation pipeline for DNA microarrays. BMC Bioinformatics. 2009;10:30. doi: 10.1186/1471-2105-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(Suppl.):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manda P, Freeman MKG, Bridges SM, Jankun-Kelly TJ, Nanduri B, McCarthy FM, Burgess SC. GOModeler- a tool for hypothesis-testing of functional genomics datasets. BMC Bioinformatics. 2010;11(Suppl. 6):S29. doi: 10.1186/1471-2105-11-S6-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders WS, Bridges SM, McCarthy FM, Nanduri B, Burgess SC. Prediction of peptides observable by mass spectrometry applied at the experimental set level. BMC Bioinformatics. 2007;8(Suppl. 7):S23. doi: 10.1186/1471-2105-8-S7-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridges SM, Magee GB, Wang N, Williams WP, Burgess SC, Nanduri B. ProtQuant: a tool for the label-free quantification of MudPIT proteomics data. BMC Bioinformatics. 2007;8(Suppl. 7):S24. doi: 10.1186/1471-2105-8-S7-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harhey G, Keele J, Smith TPL, Alexander LJ, Matukumalli LK, Schroeder SG, Liu G, Van Tassell C, Sonstegard T. Plant and Animal Genome XVI Conference, January 12–16. San Diego, CA: Town & Country Convention Center; 2008. Description and analysis of the bovine gene atlas an extensive compendium of bovine transcript profiles. Poster P516: Cattle. [Google Scholar]
- 38.Kelley R, Harper J, Bridges SM, Warbuton M, Hawkens L, Pechanova O, Peethambaran B, Luthe DS, Myloie J, Ankala A, et al. Integrated database for identifying candidate genes for Aspergillus flavus resistance in maize. BMC Bioinformatics. 2010;11:S25. doi: 10.1186/1471-2105-11-S6-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burt DW, Carre W, Fell M, Law AS, Antin PB, Maglott DR, Weber JA, Schmidt CJ, Burgess SC, McCarthy FM. The Chicken Gene Nomenclature Committee report. BMC Genomics. 2009;10(Suppl. 2):S5. doi: 10.1186/1471-2164-10-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar R, Nanduri B. HPIDB - a unified resource for host-pathogen interactions. BMC Bioinformatics. 2010;11(Suppl. 6):S16. doi: 10.1186/1471-2105-11-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference Genome Group of the Gene Ontology Consortium. The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]