Abstract
The Biological General Repository for Interaction Datasets (BioGRID) is a public database that archives and disseminates genetic and protein interaction data from model organisms and humans (http://www.thebiogrid.org). BioGRID currently holds 347 966 interactions (170 162 genetic, 177 804 protein) curated from both high-throughput data sets and individual focused studies, as derived from over 23 000 publications in the primary literature. Complete coverage of the entire literature is maintained for budding yeast (Saccharomyces cerevisiae), fission yeast (Schizosaccharomyces pombe) and thale cress (Arabidopsis thaliana), and efforts to expand curation across multiple metazoan species are underway. The BioGRID houses 48 831 human protein interactions that have been curated from 10 247 publications. Current curation drives are focused on particular areas of biology to enable insights into conserved networks and pathways that are relevant to human health. The BioGRID 3.0 web interface contains new search and display features that enable rapid queries across multiple data types and sources. An automated Interaction Management System (IMS) is used to prioritize, coordinate and track curation across international sites and projects. BioGRID provides interaction data to several model organism databases, resources such as Entrez-Gene and other interaction meta-databases. The entire BioGRID 3.0 data collection may be downloaded in multiple file formats, including PSI MI XML. Source code for BioGRID 3.0 is freely available without any restrictions.
INTRODUCTION
Cellular behavior is dictated by a complex global network of protein interactions, which are modified at the level of gene expression, messenger RNA (mRNA) stability, translation rate, degradation, localization and post-translational modifications (1). The interconnectivity of the physical interaction network is reflected by a massively dense network of genetic interactions (2). Perhaps the single most important problem in biology and biomedicine is to understand how these myriad interactions encode stable phenotypes at the cell, tissue and organism level. Recent high-throughput (HTP) approaches for comprehensive determination of protein and genetic interactions, when combined with gene expression and phenotypic profiles, afford the potential for systems-level interrogation of biological responses (3). However, while the discovery and analysis of biological interactions now underpin much of modern biomedical research, the interpretation and integration of the morass of data in the primary literature have been seriously hindered by the failure to capture this information in a distilled and consistently codified fashion (4). This shortfall is exacerbated by the dearth of reliable tools to manage, integrate and query large data sets. To help address this dilemma, we developed the BioGRID database as an open source repository for protein and genetic interactions as systematically curated from the primary biomedical literature (5). The BioGRID provides a user-friendly interface to parse individual biological interactions of interest and enables the integration of data embedded in the primary literature with HTP data sets. These federated data sets allow the construction of interaction networks from various data sources, which can be used to explain biological processes, interpret different orthogonal data types and predict new biological functions (6,7).
DATA CONTENT
The BioGRID captures, annotates and distributes comprehensive collections of physical and genetic interactions from model organisms in a standardized format based on a uniform set of experimental evidence codes. BioGRID provides full annotation support for 50 species of biomedical relevance, and currently archives interactions for Homo sapiens, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, Drosophila melanogaster, Bacillus subtilis, Bos taurus, Caenorhabditis elegans, Canis familiaris, Danio rerio, Escherichia coli, Gallus gallus, human herpesvirus, Macaca mulatta, Mus musculus, Rattus norvegicus and Xenopus laevis. Annotation resources for all supported species are routinely updated to prevent ambiguous or out-of-date nomenclature. The BioGRID houses over 17 million gene names, aliases, systematic names and external database identifiers to provide relevant search results.
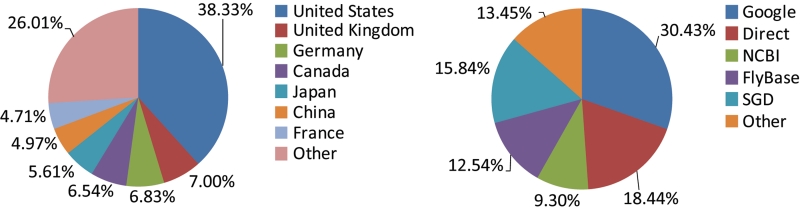
As of 1 August 2010 (version 3.0.67) BioGRID contains 347 966 (243 005 non-redundant) interactions comprised of 177 804 (123 683 non-redundant) protein and 170 162 (124 410 non-redundant) genetic interactions (Table 1). Of these curated interactions, ∼31% are drawn from 22 653 focused low-throughput (LTP) studies and 69% stem from 113 HTP studies, defined as reporting more than 100 interactions in tabular form. Through monthly updates, complete coverage of the entire literature is maintained for the budding yeast S. cerevisiae (currently 236 850 interactions) and the fission yeast S. pombe (currently 15 546 interactions). Comprehensive curation of the primary literature for protein interactions has also been completed for the model plant A. thaliana (currently 4893 interactions). These collated data sets are available from the BioGRID website for both searches and downloads, and are fully linked from the respective model organism databases Saccharomyces Genome Database (SGD, S. cerevisiae) (8), GeneDB (GDB, S. pombe) (9) and The Arabidopsis Information Resource (TAIR, A. thaliana) (10). BioGRID supports an international user base, as directed from a variety of traffic sources (Figure 1).
Table 1.
Increase in BioGRID data content since previous update
| Organism | Type | June 2006 (2.0.17) |
August 2010 (3.0.67) |
||||
|---|---|---|---|---|---|---|---|
| Nodes | Edges | Papers | Nodes | Edges | Papers | ||
| A. thaliana (thale cress) | PI | 0 | 0 | 0 | 1735 | 4719 | 747 |
| GI | 0 | 0 | 0 | 88 | 174 | 55 | |
| C. elegans (nematode worm) | PI | 2790 | 4433 | 1 | 2813 | 4663 | 12 |
| GI | 0 | 0 | 0 | 1030 | 2112 | 5 | |
| D. melanogaster (fruit fly) | PI | 6997 | 22 113 | 2 | 7396 | 24 480 | 167 |
| GI* | 1189 | 10 314 | 1493 | 982 | 9994 | 1466 | |
| H. sapiens (human) | PI | 3380 | 7238 | 178 | 9467 | 48 368 | 10 203 |
| GI | 0 | 0 | 0 | 479 | 463 | 178 | |
| S. cerevisiae (budding yeast) | PI | 5144 | 49 297 | 3267 | 5783 | 90 769 | 5444 |
| GI | 3352 | 24 636 | 3796 | 5357 | 146 081 | 5606 | |
| S. pombe (fission yeast) | PI | 0 | 0 | 0 | 1441 | 4019 | 769 |
| GI | 0 | 0 | 0 | 1340 | 11 527 | 953 | |
| All other organisms | PI | 284 | 620 | 73 | 2288 | 2985 | 830 |
| Total | ALL | 19 176 | 118 671 | 7818 | 30 665 | 347 966 | 23 451 |
Data drawn from monthly releases 2.0.17 and 3.0.67 of BioGRID.
Nodes refers to genes or proteins, edges refers to interactions.
PI, protein interactions; GI, genetic interactions.
*indicates interactions from FlyBase.
Figure 1.
Distribution of BioGRID users (left) and traffic sources (right).
Literature coverage for animal model species, including humans, is far from complete (see below). Recent curation efforts have focused on human protein interactions, given both the richness of data and the obvious biomedical relevance. A recent BioGRID curation drive has increased the number of annotated human protein interactions to 48 831 (33 017 non-redundant) as drawn from more than 10 000 publications (Table 1). As part of this effort, we have undertaken new cross-species curation initiatives in areas of chromatin remodeling (CR) and the ubiquitin proteasome system (UPS) that have thus far resulted in 12 455 (7529 non-redundant) and 6875 (4881 non-redundant) interactions, respectively. All of the interactions from these ongoing projects are updated monthly and made available on the BioGRID website for download. Analogous curation drives are being coordinated with WormBase for C. elegans (11) and the Gene Ontology (GO) Consortium for the 12 GO Reference Genomes (12).
Interaction data in the BioGRID is augmented by other associated data types for some species. Protein phosphorylation plays a critical role in controlling the dynamics of cellular interaction networks, and the recent development of mass spectrometric methods for HTP detection has resulted in a deluge of phosphorylation sites. To complement HTP data sets, we developed a resource for budding yeast phosphorylation site data captured from the primary literature, called PhosphoGRID (http://www.phosphogrid.org). This sister database currently contains 6440 experimentally verified in vivo phosphorylation sites found on 1770 proteins, as curated from 329 publications (13). In collaboration with SGD, we have also curated 126 818 phenotypes for genetic interactions in both budding and fission yeast, which are available in search results and as downloads from BioGRID and SGD (8).
CURATION PRACTICE
BioGRID captures biological interactions as binary representations, as well as n-way representations of protein complexes or contextual genetic background effects, all defined by a structured classification of experimental methods. Additional relevant attributes, including post-translational modifications, strain background and experimental conditions are also captured. Interaction data are curated from peer-reviewed publications by a team of PhD-level curators using an automated Interaction Management System (IMS). All curated interactions must be supported by published experimental evidence, either in primary figures and tables or in associated supplementary material; interactions cited in review articles, as data not shown or as personal communications are not curated. A list of BioGRID evidence codes and a mapping to the PSI-MI ontology (14) can be found on the BioGRID wiki (http://wiki.thebiogrid.org) page.
A major upgrade to the IMS supports rigorous curation procedures across multiple international sites. New features in IMS 2.0 include: (i) enhanced quality control via an automated gene/protein verification system; (ii) multiple simultaneous curator access and project customization; (iii) support for annotation of phenotypes, post-translational modifications, complexes and other non-pairwise interactions; (iv) error detection linked directly to MOD partners; (v) automated and prioritized publication queues based on text mining approaches or gene/protein lists from partner databases; (vi) enhanced quality control and history tracking of all curated interactions; (vii) an improved AJAX-based curation interface; and (viii) HTP data set upload tools with built-in error detection. The IMS 2.0 architecture has been migrated to PHP and elaborated to support interaction record tags for assignment of specific datasets and/or data types.
CURATION STRATEGIES
Systematic curation of the entire biomedical literature is obviously an enormous task (4). For instance, PubMed contains well over 11 million publication entries for human alone, and even pathogen-specific publications can number in the hundreds of thousands. While complete interaction coverage has been obtained for the two main model yeasts, and may be achievable for metazoan model organisms, this purely unbiased approach is not practical for manual curation of human literature. An effective alternative is to undertake focused curation drives in more restricted realms in order to encapsulate interaction data in specific biological processes, human diseases or conserved networks. This approach has several benefits including the development of curator expertise in particular areas, which results in improved curation throughput and accuracy, and the capacity to focus on biological networks of relevance in drug discovery. Additionally, themed curation projects can be initiated in collaboration with experts in their respective domain to develop up-to-date, comprehensive and reliable data sets. Thematic curation projects within BioGRID have included target of rapamycin (TOR), TGF-β and breast cancer signaling pathways, as well as the conserved CR and UPS networks noted above. A cross-species curation drive on the Wnt pathway, which is being comprehensively annotated by the GO Reference Genome Project (12), is underway by BioGRID and WormBase curators. These focused data sets can be retrieved as download files from the BioGRID website.
Previously curated publications annotated in other resources can serve as an effective guide to expedite curation of interaction data. The GO Consortium annotates genes/proteins to biological processes, molecular function and cellular localization on the basis of various experimental and contextual support, including inferred from protein interaction (IPI) or inferred from genetic interaction (IGI) evidence codes (12). GO annotations have been used to infer biological interaction networks (15). BioGRID uses IPI and IGI evidence codes to facilitate extraction of experimental data that support interactions for a given gene/protein. For example, a systematic effort to extract interaction data from all C. elegans publications annotated by GO is currently in progress by BioGRID and WormBase curators (11). Similarly, in conjunction with MOD curators, lists of priority publications likely to contain interaction data based on keyword searches are parsed from MOD gene/protein records.
The BioGRID curation process is enabled by text mining of the literature to identify publications that are more likely to contain biological interaction data. Text-mining approaches have been developed that enrich for publications with specific data attributes, including Textpresso (16) and iHOP (17). NCBI MeSH terms and simple Boolean queries on PubMed also help to prioritize publication lists for curation. While text-mining approaches serve to prioritize publication lists for BioGRID curators, these approaches are currently not definitive. Direct comparisons between text mining and exhaustive manual curation reveal substantial false positive and false negative error rates (18). In order to contribute to the development of text-mining strategies, BioGRID has recently participated in the BioCreative challenge, a community-wide effort to evaluate text mining and information extraction systems in biomedicine (18). In collaboration with the MINT interaction database (19), BioGRID has provided gold standard manually curated datasets and expert knowledge for various classification and extraction tasks in the BioCreative III Challenge. These benchmarks will enable the refinement of various text-mining approaches (see http://www.biocreative.org for details).
Direct author deposition represents a final important source of interaction data. The BioGRID processes large pre-publication data sets provided by authors for immediate release upon the publication date (2,7,20). Submissions can be coordinated through the BioGRID curation team using templated Microsoft Excel or plain text submission forms that are available on the BioGRID wiki page (http://wiki.thebiogrid.org). BioGRID curators will assist VMWare-based authors in data compilation and implementation of correct record structures. In addition to large data sets from HTP studies, author-directed curation represents an untapped source of interaction data for the entire biomedical literature. A significant fraction of curator time is spent on parsing publications with few or no interactions and on correctly assigning gene identifiers (31). The International Society for Biocuration has advocated that a standardized author form be used to document biological interactions contained in each publication in association with the publication process, much as authors are currently required to provide Genbank, PDB and microarray accession numbers prior to publication. If implemented, this process would greatly improve curation coverage and liberate curators to assure curation quality (4).
DATA DISSEMINATION
All interaction records in BioGRID can be freely downloaded in a wide variety of standard formats, including PSI-MI (14) and tab-delimited text files. BioGRID data are also available through partner model organism databases (MODs) including SGD (8), FlyBase (21), TAIR (10) and GeneDB (9), through external resources such as NCBI Entrez-Gene, DroID (22) and GermOnline (23), and through meta-databases such as STRING (24), iRefIndex (25) and Pathway Commons (http://www.pathwaycommons.org). BioGRID interaction data can also be accessed automatically through biological network visualization applications that include Osprey (26), Cytoscape (27), GeneMania (28) and ProHits (29).
BioGRID participates in a number of international initiatives that aim to provide the biomedical community with molecular interaction data, phenotypic descriptions of gene function and model organism annotation. BioGRID is a member of the IMEx consortium (http://www.imexconsortium.org), which coordinates the curation of interaction data according to the PSI-MI standard for dissemination across all partner databases (30). BioGRID actively collaborates with MODs on different aspects of curation. For example, in collaboration with SGD and GeneDB, BioGRID curators have assigned structured phenotypes to over 126 000 yeast genetic interactions. BioGRID has begun to coordinate its curation activities with the Linking Animal Models to Human Disease Initiative (LAMHDI) in order to build and predict networks of interactions associated with human disease (http://www.lamhdi.org).
USER INTERFACE IMPROVEMENTS
The BioGRID 3.0 web interface has been redesigned to allow cross-species searches from a single unified home page. BioGRID searches can be performed by gene/protein identifier, PubMed identifier or publication keywords. The new BioGRID search result page is enhanced with features including tooltips, pagination and animated transitions for detailed record attributes, such as quantitative interaction scores (Figure 2). Interaction data views can be toggled between a sortable table and a collapsed summary layout. Data types can also be filtered between HTP, LTP or combined data. The BioGRID download page has been converted to an AJAX-based engine for on-the-fly creation of interaction files for any gene/protein or curated publication. Larger interaction datasets can be selected by experimental evidence code, publication, species or database version number. A custom data set tool enables interactions from several publications and/or gene/proteins to be merged into a single download file. All results can be downloaded in IMEx-compatible PSI-MI 2.5.3 XML and PSI-MI Tab formats (14), as well as tab-delimited text files that are compatible with visualization software (26–28). BioGRID documentation has been migrated to a wiki-based platform that facilitates maintenance and quality assurance. Online documentation now covers evidence codes, download formats, interpretation of search results and custom data set construction. BioGRID interaction statistics have been upgraded to reflect monthly updates and specific values for each search result page.
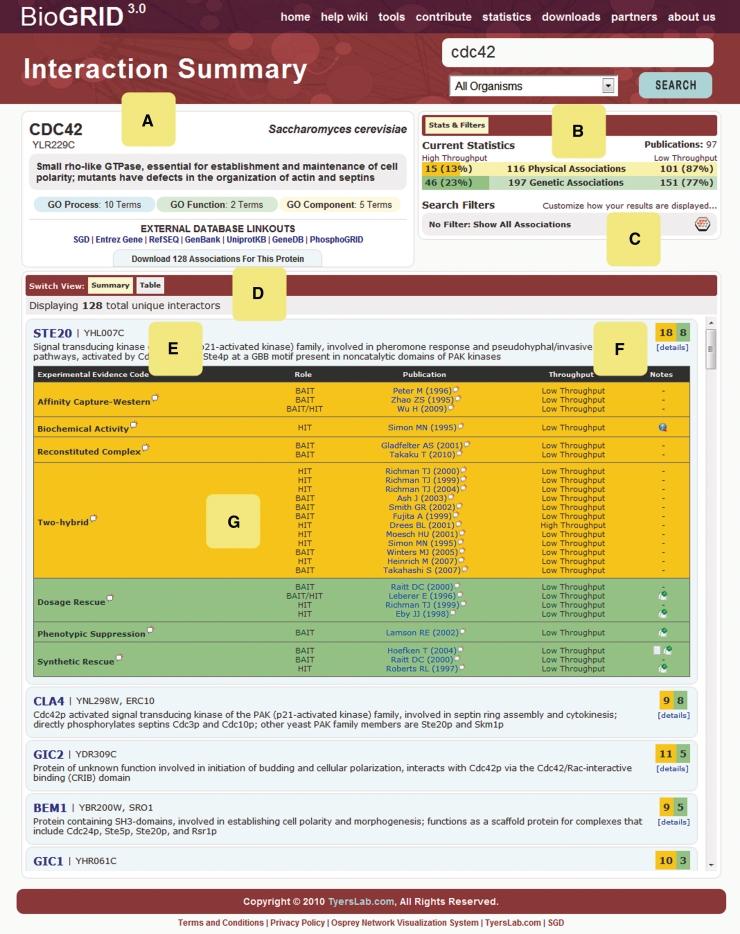
Figure 2.
Sample BioGRID 3.0 search result layout. (A) Search annotation includes description of gene/protein function, gene aliases, external database linkouts and Gene Ontology (GO) annotation. (B) Statistics for physical and genetic interactions, subdivided into HTP and LTP data. (C) Search filters enable user-customized display of results. (D) Interaction view options. (E) Annotation for each interaction partner. (F) Summary of protein and genetic interaction replicates. (G) Interaction details for publication, role as bait or hit, experimental evidence code, data type and applicable free text notes.
DATABASE IMPROVEMENTS
The BioGRID database architecture has been redesigned and migrated to a fault-tolerant VMWare-based hardware architecture that provides greater flexibility and redundancy against failure. BioGRID 3.0 supports ∼17 million systematic names, aliases, official symbols and external identifiers from Ensembl, UniprotKB, NCBI Refseq, Entrez-Gene, Genbank, SGD, WormBase, FlyBase, MGD and TAIR, amongst other sources. Compared to the previous release (5), BioGRID 3.0 supports annotation for an additional 37 organisms, including viruses and bacterial species. The annotation update cycle time has been markedly improved such that a complete update can be performed in 2 or 3 days. Frequent annotation updates improve search engine speed, ensure search result accuracy and avoid the common nomenclature pitfalls of ambiguous and/or deprecated identifiers. BioGRID search results are prioritized by official symbols and systematic names in rank order of number of interactions. The BioGRID website and download files are updated each month according to a simplified and stable update procedure. MOD partners draw selected datasets from BioGRID on a similar monthly schedule. As with previous versions, source code for BioGRID is freely available without restrictions.
FUTURE DEVELOPMENTS
BioGRID curation will continue to emphasize major model organism species, with the goal of achieving comprehensive literature coverage for S. cerevisiae, S. pombe, A. thaliana, C. elegans and D. melanogaster. Curation drives for human interactions will be focused in part on specific biological processes, including chromatin remodeling, the ubiquitin proteasome system and phosphorylation-based signaling. In collaboration with LAMHDI, curation will be extended to specific human disease processes, and will include annotation of disease gene-phenotype relationships. Identification of relevant publications for disease gene curation will be facilitated by text-mining approaches. The next version of the BioGRID website will improve the user interface by providing user login accounts, themed interaction record tags, custom search filters and web-based visualization tools. BioGRID data will also be made readily available to other third-party applications through web services. Through these approaches, the BioGRID will continue to capture and codify biological interaction data for the biomedical research community.
FUNDING
The National Institutes of Health National Center for Research Resources (1R01RR024031 to M.T. and K.D.); the Biotechnology and Biological Sciences Research Council (BB/F010486/1 to M.T.); the Canadian Institutes of Health Research (FRN 82940 to M.T.); the European Commission FP7 Program (2007-223411 to M.T.); the NIH National Human Genome Research Institute (P41-HG02223 to WormBase); a Royal Society Wolfson Research Merit Award (to M.T.); and the Scottish Universities Life Sciences Alliance through the Scottish Funding Council (to M.T.). Funding for open access charges: National Institutes of Health (1R01RR024031).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Mike Cherry, Paul Sternberg, Bill Gelbart, Gary Bader, Quaid Morris, Olga Troyanskaya, David Botstein, Henning Hermjakob, Shoshana Wodak, Ian Donaldson, Anne-Claude Gingras, Val Wood, Jurg Bahler, Lincoln Stein and Judy Blake for support and helpful discussions.
REFERENCES
- 1.Bork P, Jensen LJ, von Mering C, Ramani AK, Lee I, Marcotte EM. Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat. Rev. Genet. 11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, et al. Big data: the future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huttenhower C, Mutungu KT, Indik N, Yang W, Schroeder M, Forman JJ, Troyanskaya OG, Coller HA. Detailing regulatory networks through large scale data integration. Bioinformatics. 2009;25:3267–3274. doi: 10.1093/bioinformatics/btp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces genome database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudet P, Chisholm R, Berardini TZ, Dimmer E, Engel SR, Fey P, Hill D, Howe D, Hu JC, Huntley R, et al. The gene ontology's reference genome project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark C, Su T-C, Breitkreutz A, Lourenco P, Dahabieh M, Breitkreutz B-J, Tyers M, Sadowski I. PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database: The Journal of Biological Databases and Curation. 2010 doi: 10.1093/database/bap026. doi:10.1093/database/bap026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drabkin HJ, Hollenbeck C, Hill DP, Blake JA. Ontological visualization of protein–protein interactions. BMC Bioinformatics. 2005;6:29. doi: 10.1186/1471-2105-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez JM, Hoffmann R, Valencia A. iHOP web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leitner F, Mardis SA, Krallinger M, Cesareni G, Hirschman LA, Valencia A. An overview of BioCreative II.5. IEEE/ACM Trans. Comput. Biol. Bioinform. 2010;7:385–399. doi: 10.1109/tcbb.2010.61. [DOI] [PubMed] [Google Scholar]
- 19.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Pacifico S, Liu G, Finley RL., Jr DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiederkehr C, Basavaraj R, Sarrauste de Menthiere C, Hermida L, Koch R, Schlecht U, Amon A, Brachat S, Breitenbach M, Briza P, et al. GermOnline, a cross-species community knowledgebase on germ cell differentiation. Nucleic Acids Res. 2004;32:D560–D567. doi: 10.1093/nar/gkh055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(Suppl.):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, Zhang JP, Larsen B, Stark C, Breitkreutz A, Lin Z-Y, Breitkreutz B-J, Ding Y, Colwill K, Pasculescu A, et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 2010;10:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orchard S, Aranda B, Hermjakob H. The publication and database deposition of molecular interaction data. Curr. Protoc. Protein Sci. 2010 doi: 10.1002/0471140864.ps2503s60. Chapter 25, Unit 25.3. [DOI] [PubMed] [Google Scholar]
- 31.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimal information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;8:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]