Abstract
The allele frequency net database (http://www.allelefrequencies.net) is an online repository that contains information on the frequencies of immune genes and their corresponding alleles in different populations. The extensive variability observed in genes and alleles related to the immune system response and its significance in transplantation, disease association studies and diversity in populations led to the development of this electronic resource. At present, the system contains data from 1133 populations in 608 813 individuals on the frequency of genes from different polymorphic regions such as human leukocyte antigens, killer-cell immunoglobulin-like receptors, major histocompatibility complex Class I chain-related genes and a number of cytokine gene polymorphisms. The project was designed to create a central source for the storage of frequency data and provide individuals with a set of bioinformatics tools to analyze the occurrence of these variants in worldwide populations. The resource has been used in a wide variety of contexts, including clinical applications (histocompatibility, immunology, epidemiology and pharmacogenetics) and population genetics. Demographic information, frequency data and searching tools can be freely accessed through the website.
INTRODUCTION
Over recent years, a vast number of studies investigating the incidence of several genes involved in the immune system response have been reported in the literature. The extensive number of DNA sequence variants, differences in frequencies found among populations and their importance in scientific and clinical areas led to the implementation of a central source to store these frequencies (1).
The allele frequency net database (AFND) is a public electronic resource dedicated to the storage of allele, haplotype and genotype frequencies of several immune genes. The information available on this database consists of genes principally related to the major histocompatibility complex (MHC). In humans, one of the main components of this complex is the human leukocyte antigen (HLA) system, which is considered to be the most polymorphic region in the human genome. With more than 100 genes suggested to have immunological functions (2), this genomic region has been studied for many years with notable advances in clinical immunology (3). The HLA system is recognized for its importance in transplantation (4), associations in infectious diseases (5), autoimmune diseases (6) and studies of diversity in populations (7).
The AFND also contains information about other immune genes such as the killer-cell immunoglobulin-like receptors (KIR). Present on natural killer cells (NK), the products of some of these KIR genes interact with HLA Class I molecules and are suggested to influence resistance to infections, propensity to autoimmune diseases, and favorable outcome in hematopoietic stem cell transplantation (HSCT) (8,9). Additionally, the database has been expanded to contain MHC Class I chain-related (MIC) genes, which are located on the same region as HLA genes and associated to some diseases and rejection in transplantation (10). Several cytokine gene polymorphisms are also included, which are proteins secreted in the immune response and linked to a number of diseases (11).
The goal of the AFND is to serve as a warehouse of frequency data sets and provide an online repository with a set of querying tools for the examination of frequencies in different populations. Since the initial version of the website, a number of tools have been incorporated to improve searching mechanisms and include other polymorphisms. With more than 4000 allelic variants described in the IMGT/HLA database (12) as of Release 3.0.0 April 2010, the AFND constitutes an up-to-date data source for the examination of frequencies and confirmation of presence of HLA alleles. The web-based framework provides individuals with an online submission system, allowing data to be contributed by the wider research community. One of the main features that distinguishes the AFND is that data sets stored have been manually curated, through a process of data validation to provide researchers with accurate results. Data and searching tools can be freely accessed by any individual through the http://www.allelefrequencies.net portal.
IMPLEMENTATION
To facilitate the maintenance and retrieval of information, the back-end of the AFND is based on a relational database model utilizing MySQL as the database management system. The database can be accessed utilizing any of the most common web browsers. The use of a web browser as a front-end gives the facility to users to access data without the necessity of installing a package. Web pages were implemented using the active server pages scripting environment for the development of dynamic pages, with the assistance of the JavaScript language for data entry validation. Additionally, the asynchronous JavaScript and XML (AJAX) technology was included in some of the searching mechanisms to allow simpler user interaction and improved visualizations.
The graphical display of this software was developed using HTML and CSS to guarantee a standard visualization in the majority of common browsers. Presently, all web pages have been tested on Internet Explorer 8®, Firefox 3.5®, Safari 4®, Opera 9® and Google Chrome 4®.
WEBSITE ORGANIZATION AND CONTENT
The allele frequency net website is divided into four main sections: HLA, KIR, MIC and cytokine frequencies. Each section consists of different querying tools depending on the availability of data in each polymorphic region, i.e. haplotype, genotype or allele frequency, breakdowns to summarize the existing data in each polymorphism and other online tools such as searches for rare HLA alleles and the frequency of particular amino acids within a given position in a population (Table 1). Searches in each section have been designed with a set of instructions on how to perform the query. The AFND is regularly updated to include new user submissions and information from relevant peer-reviewed publications. Additionally, due to the constant increase in the number of alleles identified by molecular methods, the database is periodically updated according to the official nomenclature from latest releases available on the IMGT/HLA and IPD-KIR databases (12,13). At present, alleles on the website have been updated containing the most recent nomenclature guidelines for allele designations (14).
Table 1.
Overview of the main searches and tools available at the www.allelefrequencies.net website
| Polymorphic region | Tool | Description |
|---|---|---|
| HLA | AFS | Querying tool to explore allele and phenotype frequencies in one or many populations from the given criteria |
| Haplotype frequency search | Tool to consult a particular haplotype in a set of populations at two or more loci. | |
| Rare HLA alleles search | Section to ascertain the rarity of HLA alleles based on the confirmation from different databases and individual laboratories | |
| Rare HLA allele detector | Tool to detect rare and very rare alleles from large data sets | |
| Amino acid frequencies in populations | Interactive tool to examine frequencies in a given population comparing alleles differing at amino acid positions | |
| KIR | AFS | Querying tool to explore allele and phenotype frequencies in one or many populations from the given criteria |
| Genotype frequency search | Tool which allows users to display genotype frequencies of a given profile | |
| Cell lines and CEPH families | Section that provides a reference of KIR cell-lines and CEPH families typed at allele level | |
| Cytokines | Cytokine frequency search | Querying tool to explore allele frequencies in one or many populations from the given criteria |
| MIC | AFS | Querying tool to explore allele and phenotype frequencies in one or many populations from the given criteria. |
| MIC–HLA association frequency search | Section that provides frequencies of MIC and HLA associations | |
| Online submissions | Populations | Online submission tool for a new population |
| Confirmation of rare alleles | Section for the confirmation of rare HLA alleles |
Population data sets
The collection of populations available on the AFND consists of 1133 population samples from 608 813 healthy unrelated individuals. The compilation comprises more than 100 000 records at allele, haplotype and genotype level within which HLA comprises 90% of the allele frequency entries (Table 2). These populations are divided in 786 HLA populations, 181 KIR populations, 110 Cytokines populations and 56 MIC populations. Populations available on the database are mainly derived from peer-reviewed publications or from direct submissions to the website from individual laboratories. Our aim is to capture all previously published studies (between 1990 to present) and we believe that the vast majority of published data sets have been included in AFND. To date, this includes publications from more than 65 journals (a complete list of data sets and journals can be consulted via http://www.allelefrequencies.net/datasets.asp). Based on the interest of the user, the data may be searched according to the source (i.e. published | sent direct; anthropology studies, etc.). The bibliographic reference for each study is provided so that a user may verify what type of analysis the author has used for calculating frequencies. Frequency data submitted directly by an individual has typically been obtained by direct counting. When data has been published or sent directly to the AFND in which the author has not been able to differentiate some alleles (i.e. ambiguous data), a note in the publication details is used to describe how the frequency data was entered for one of the ambiguous alleles. For example, ‘unable to differentiate alleles that are identical over exons 2 and 3 (Class I) or identical over exon 2 (Class II)’, and where frequencies are reported for the first allele only.
Table 2.
Frequency data sets by polymorphic region at AFND
| Frequencies |
|||||
|---|---|---|---|---|---|
| Polymorphic Region | Populations | Individuals | Allele | Haplotype | Genotype |
| HLA | 786 | 562 689 | 81 160 | 7426 | – |
| KIR | 181 | 20 965 | 4620 | – | 2398 |
| Cytokine | 110 | 17 864 | 3555 | – | – |
| MIC | 56 | 7295 | 678 | 244 | – |
| Total | 1133 | 608 813 | 90 013 | 7670 | 2398 |
Submission of data
One of the most important objectives in the design of the website was to provide users with an online submission form to incorporate their own studies through the website and which ensures consistency of demographic data. To do this, individuals are assisted during the submission process with drop down boxes to provide basic information related to demographic data such as a descriptive name of the population (country name, geographic region and ethnic origin), sample size, polymorphic region, latitude and longitude coordinates (if known), family background, methods used in typing and references in literature, if the study has been published. If a publication uses an ethnicity code that is not included in the drop down box, the ethnicity given by the submitter is added to the current list As such, a list of ethnicity codes is maintained in AFND to standardize reporting, although as yet we do not map these to any wider community controlled vocabulary (see ‘Discussion’ section). Individuals are requested to input the corresponding frequencies through an online web form or by providing a pre-formatted spreadsheet containing frequency results. User submissions then undergo a data validation procedure, performed by a group of curators of the AFND. Some of the validations include the selection of an appropriate name of the population to best describe the origin, i.e. Name of country followed by region and ethnicity if known. If a population submitted by a second group of individuals is geographically and ethnically similar to an existing population on the database, a consecutive number is assigned to that population to differentiate both data sets and to allow them to be compared (e.g. China Guangzhou Han, China Guangzhou Han pop 2). Therefore, the system has been designed to validate duplications. Other controls performed include verifying that the correct and current nomenclature has been used for an allele and, if not, the allele name is updated. The database contains the current definition of alleles, thus, data entered directly to the website will contain correct allele names. If necessary, the author of the data is contacted with any query or any change made by curators. For frequency data, values are added and for any summation greater or less than 1 the author is contacted. If there are frequencies that are >1 which cannot be explained, the submission is rejected. Frequencies which sum <1 are kept in the database. Unfortunately, on many occasions, data that is published is not always correct and editors of journals concerned are contacted to discuss these issues. Whilst the AFND cannot assess the typing accuracy of data provided, >90% of the data on the website has been peer-reviewed and published. Thus, the AFND relies on the accuracy of data being verified by the reviewers of the journals and acts mainly as a source for compiling data. It is our intention in the future to collect the raw data in order that we can be more proactive in assessing the quality of data.
Allele frequency searches
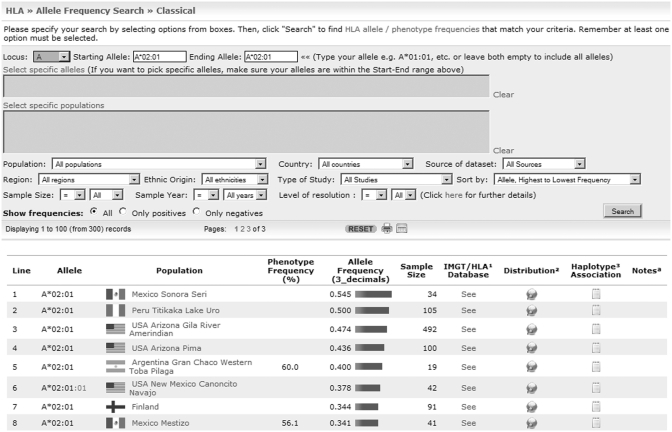
The most commonly used tool within AFND is the allele frequency search (AFS), with which users can examine the frequency of a particular allele in the existing population data sets, by filtering results with a set of criteria. The AFS is available for all polymorphisms on the website. To perform the search, users usually start with the selection of a locus and a particular allele to identify which populations are more likely to present the allele. To extend the searching criteria, users can select one, several or all populations, a set or range of alleles, country, geographic region, ethnicity and/or the year in which data was submitted (Figure 1). In HLA, MIC and KIR polymorphisms, alleles can be typed at different levels of resolution (i.e. allele group, specific HLA protein, synonymous allele with a substitution within the coding region and differences in a non-coding region in that order, e.g. HLA-A*01:01:01:01) (14,15). The official nomenclature available on the IMGT/HLA and IPD-KIR databases describes alleles only at the highest resolution. To ensure that high resolution data can be retrieved when a low level resolution allele is selected, the search uses parsing methods to display all information that may be relevant to the user. For instance, a search for the HLA-A*02:01 allele will also display incidences of alleles at high resolution that start with HLA-A*02:01. Additionally, users are able to optimize their queries to further refine data sets by selecting populations with a sample size from a range of values and/or a specific level of resolution. Populations from recent years are more likely to contain alleles with a high resolution level and thus, more accurate data. Furthermore, recent additions include filters to search information on a specific source of data set and type of study, for example populations available in the literature oriented to anthropology studies. Results displayed in the search include the allele name, name of the population, allele and/or phenotype frequency and the sample size of the population to estimate the number of individuals who carry the allele. By clicking on the ‘Population Name’ hyperlink users can access demographic details of the population in which the allele is present. The list of output records can be sorted by allele or population and the corresponding frequency from highest to lowest value. Also, haplotype associations and graphical distribution overlaid on world maps are some of the recent options added for each record.
Figure 1.
Screen shot of the HLA AFS. The figure shows an example of a search of the HLA-A*02:01 allele sorted by highest to lowest frequencies. Other data provided includes a link to the IMGT/HLA database for sequence information of the allele, link to frequency distribution maps, haplotype association of the allele and additional notes. The information provided can be printed to include all records.
Haplotype frequency search
Following a similar multiple filter scheme, the AFND repository also includes a tool for querying haplotype frequencies from 7426 HLA haplotypes and 244 MICA-HLA-B association records from 147 325 individuals. At present, the collection of haplotypes consists of 344 globally distributed populations in 79 countries. The program permits the user to customize a frequency search by inputting an allele for one or more loci and search for associated haplotypes. Results can be filtered by a particular population, country, source of data, geographic region, ethnicity of the individual and number of loci tested for the haplotype. The haplotypic information can be more useful than information only on the allele, especially in clinical applications. Therefore, this search can be used as a complement of haplotype searches performed in bone marrow and solid organ transplant registries in which, on some occasions, the information about the ethnicity of the individual is unknown. Haplotypes can also be searched at lower or higher resolution and from two to eight routinely typed HLA loci (HLA-A, -B, -C, -DRB1, -DPA1, -DPB1, -DQA1 and -DQB1).
Genotype frequency search
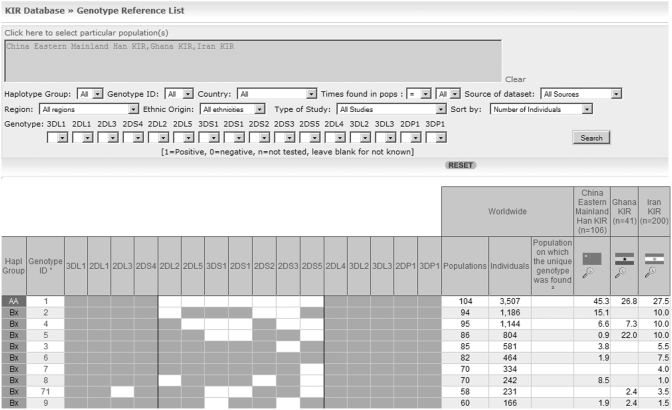
One of the most recent developments included in the website has been the compilation of an inventory of KIR genotype profiles published in the literature. This section comprehends the most extensive archive of KIR genotypes and their corresponding frequencies in worldwide populations. Presently, the genotype data encompasses 2398 records of which 368 distinct KIR genotype profiles have been identified. The KIR genotype composition consists of 16 genes, which may be present or absent in a specific genotype. In the system, users can search for a particular genotype and examine its corresponding frequency from a list of 102 KIR populations available with genotype data. The genotype search provides different approaches to find the incidence of a specific profile. A list of all genotypes and the number of populations and individuals in which the profile has been found appears on the main screen. Users are provided with a range of options including the selection of one or specific populations and one or many genes that constitute the genotype. The information displayed after performing the search comprises the genotype, the id of the genotype, which is automatically assigned by the AFND as a consecutive number, the haplotypes AA, Bx (where x can be A or B) which constitutes the genotype and the genotype frequency of populations considered on the selection (Figure 2). If the genotype is not found from the initial search, users are provided with a list of the closest genotypes differing by one gene.
Figure 2.
Screen shot of the KIR genotype frequency search. The figure displays a view of the first 10 genotypes found in three populations (China Eastern Mainland Han, Ghana and Iran) sorted by the number of individuals on which the genotype has been reported in worldwide populations.
Other online tools
Amino acid analysis
The website provides a range of tools for other analyses, including the comparison of the existing populations at amino acid level for HLA populations. One of the approaches commonly used in disease association studies is to compare frequencies of alleles between patient and control groups. We have thus developed a tool that allows users to investigate potential molecular mechanisms, by analyzing the main differences in frequencies for a specific position of the allele at the amino acid level. A summary of frequencies for each differing amino acid is presented, allowing users to compare incidences that may be implicated in the association. In the system, users can enter their own data in a tab-separated text file or select an existing population in the database. Populations and data sets provided by users must be typed at protein level (e.g. A*01:01) to be able to perform the analysis.
Rare alleles
Following the continuation of a project of the 15th International Histocompatibility Workshop (IHWS) related to the rarity of specific HLA alleles, a utility has been built to allow users to search for a particular allele and display the number of confirmations submitted by different data sources [AFND, IMGT/HLA, national marrow donor program (NMDP) in the US and individual laboratories] (16). A default mechanism uses criteria to classify the rarity of the alleles. However, the tool also allows individuals to decide whether an allele is considered to be rare by selecting their own criteria. In this search, users are invited to confirm an allele, which has been seen in their laboratories by providing basic information concerning the rare allele.
External access
An important feature of the portal is the availability of bidirectional links to different databases for data sharing and referencing. For example, in the AFS a complete list of all populations possessing the A*02:01 allele can be accessed using the http://www.allelefrequencies.net/hla6006a.asp?hla_selection=A*02:01 link, which could thus be implemented in other resources to link into the AFND. A complete list of reference links can be consulted on the ‘External access’ section of the website (http://www.allelefrequencies.net/extaccess.asp). The database maintains an active collaboration with other databases such as IMGT/HLA, IPD-KIR and NMDP for the update of nomenclature factors and confirmations of rare HLA alleles, respectively.
The site provides the option to export data to different format files including XML, tab-separated and comma-separated text files for the ‘HLA Rare Allele’ section, allowing users to integrate the information available in AFND with alternative bioinformatic packages. At present, users can print results from all searches using the printer-friendly version available for each search which can be used to export data sets in tabular format. To complement frequency data in searches for further analyses, the printer-friendly option includes information of latitude and longitude if users wish to plot frequencies on maps. Further download options will be developed in the future, in consultation with database users on their requirements.
Database users
The AFND has been extensively used in a wide range of contexts including clinical applications (Histocompatibility, Immunology, Epidemiology, Pharmacology, Rheumatology, etc.), Academic Research Centers, Research Centers (Cancer, HIV, Bone Marrow Transplant, Genomics, etc.), biotechnology and population genetics. The role of these users varies depending on their interest and is mainly categorized in three types: (i) users performing individual allele/gene frequencies queries to investigate whether an allele or haplotype from a tissue type of an individual may be frequent in a particular population, (ii) specialized users performing genetic population analysis by comparing specific frequency data sets from a particular group of populations and (iii) third-party application/database users interacting with the website by using bidirectional links and data sharing. The AFND has provided a significant resource for several genetics and cell function analyses, for example, several of the frequency data sets were used for analysis of balancing selection and heterogeneity in the HLA genes (17), characterization of populations across regions (18) and many others.
DISCUSSION
By providing an immunogenetic frequency database accessible via a web-based interface, we aim to provide the medical and scientific community with a practical approach to investigate the incidence of genes and their alleles among populations. The multiple filter schemes performed in each of the frequency searches should allow users to optimize their searches and obtain the closest matching results.
The design of the database permits the addition of other polymorphisms of interest. Ongoing developments include a new section to display frequencies of minor histocompatibility antigens (mHags) which are also of relevance in HSCT and the improvement of maps showing frequencies in different populations. In the future, we would also like to incorporate further data / statistical routines into the AFND to allow users to apply advanced bespoke analyses of whichever populations they are interested in, and subsequently select which populations to include in a map display. We also plan to include other sections in the website such as KIR gene frequencies correlated with their HLA ligand frequencies [continuing the KIR anthropology component of the 15th IHWS (19)] and a section for KIR and disease studies. We believe that several procedures on the website could be invaluable to users and we intend to provide a detailed user manual in the near future.
The AFND project also involves an active participation with international organizations, such as the HLA European Network (HLA-NET) and the Immunogenomics Data-Analysis Working Group (IDAWG) for the standardization and validation of frequency data sets, controlled vocabulary (e.g. ethnic origins, geographic regions, etc.), database schemas and data exchange.
CONCLUSION
AFND provides an important resource for the histocompatibility and immunogenetics community. Acting as a primary source, the database contains the most extensive archive of immune gene frequencies. Additionally, the development of novel mechanisms of querying and the incorporation of new polymorphisms have enriched the examination of the genes available. The database is still under active development and improvements can be applied in response to feature requests. Therefore, feedback from users is actively encouraged and will benefit the database. Full details of searches and data sets can be explored by accessing the www.allelefrequencies.net website.
FUNDING
The AFND was supported by the National Council on Science and Technology of Mexico [197198 to F.G.G.]. Funding for open access charges: Biotechnology and Biological Sciences Research Council's support for the bioinformatics group at Liverpool [BB/G010781/1 to A.R.J.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to acknowledge the support of the following sponsors whose contribution has made this project feasible: Abbott, BAG Health Care, Biotest, Gen-Probe Innogenetics, Invitrogen, Olerup SSP and One Lambda and to Ralph Komerofsky who produced the technical aspects of the first version of the website. We genuinely thank reviewers of this manuscript who have been instrumental in providing ideas for future use and development of the database.
APPENDIX 1
Access and contact
AFND Homepage: http://www.allelefrequencies.net
Contact: support@allelefrequencies.net
REFERENCES
- 1.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database. http://www.allelefrequencies.net. Tissue Antigens. 2003;61:403–407. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 2.The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 3.Thorsby E. A short history of HLA. Tissue Antigens. 2009;74:101–116. doi: 10.1111/j.1399-0039.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- 4.Claas FHJ, Duquesnoy RJ. The polymorphic alloimmune response in clinical transplantation. Curr. Opin. Immunol. 2008;20:566–567. doi: 10.1016/j.coi.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin. Microbiol. Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorsby E, Lie BA. HLA associated genetic predisposition to autoimmune diseases: genes involved and possible mechanisms. Transplant Immunol. 2005;14:175–182. doi: 10.1016/j.trim.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 8.Norman PJ, Parham P. Complex interactions: The immunogenetics of human leukocyte antigen and killer cell immunoglobulin-like receptors. Semin. Hematol. 2005;42:65–75. doi: 10.1053/j.seminhematol.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Parham P. MHC class I molecules and kirs in human history, health and survival. Nat. Rev. Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 10.Collins RWM. Human MHC class I chain related (MIC) genes: their biological function and relevance to disease and transplantation. Eur. J. Immunogenet. 2004;31:105–114. doi: 10.1111/j.1365-2370.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- 11.Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, Oksenberg J, McNicholl J, Pociot F, Hardt C, et al. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun. 1999;1:3–19. doi: 10.1038/sj.gene.6363645. [DOI] [PubMed] [Google Scholar]
- 12.Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SGE. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson J, Waller MJ, Stoehr P, Marsh SGE. IPD––the immuno polymorphism database. Nucleic Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernández-Viña M, Geraghty DE, Holdsworth R, Hurley CK, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh SGE, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens. 2003;62:79–86. doi: 10.1034/j.1399-0039.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 16.Middleton D, Gonzalez F, Fernandez-Vina M, Tiercy J-M, Marsh SGE, Aubrey M, Bicalho MG, Canossi A, Carter V, Cate S, et al. A bioinformatics approach to ascertaining the rarity of HLA alleles. Tissue Antigens. 2009;74:480–485. doi: 10.1111/j.1399-0039.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 17.Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A, Thomson G. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guinan KJ, Cunningham RT, Meenagh A, Dring MM, Middleton D, Gardiner CM. Receptor systems controlling natural killer cell function are genetically stratified in Europe. Genes Immun. 2010;11:67–78. doi: 10.1038/gene.2009.60. [DOI] [PubMed] [Google Scholar]
- 19.Hollenbach JA, Meenagh A, Sleator C, Alaez C, Bengoche M, Canossi A, Contreras G, Creary L, Evseeva I, Gorodezky C, et al. Report from the killer immunoglobulin-like receptor (KIR) anthropology component of the 15th International Histocompatibility Workshop: worldwide variation in the KIR loci and further evidence for the co-evolution of KIR and HLA. Tissue Antigens. 2010;76:9–17. doi: 10.1111/j.1399-0039.2010.01459.x. [DOI] [PubMed] [Google Scholar]