Abstract
The Rfam database aims to catalogue non-coding RNAs through the use of sequence alignments and statistical profile models known as covariance models. In this contribution, we discuss the pros and cons of using the online encyclopedia, Wikipedia, as a source of community-derived annotation. We discuss the addition of groupings of related RNA families into clans and new developments to the website. Rfam is available on the Web at http://rfam.sanger.ac.uk.
INTRODUCTION
The Rfam database maintains alignments, consensus secondary structures, covariance models (CMs) and corresponding annotation for RNA families. Each family represents a set of RNA sequences that function at the RNA level and share a clear common ancestor. Some examples are tRNA, microRNAs, spliceosomal RNAs, riboswitches, CRISPR elements and thermosensors. The primary purpose of the Rfam database is the automated, accurate annotation of non-coding RNAs (ncRNAs) in genomic sequences. Rfam is also frequently used as a source of high-quality alignments for training and benchmarking RNA sequence analysis software tools (1–5). Additionally, in the absence of a well-curated and up-to-date general RNA sequence database, equivalent to UniProt in the protein coding world, Rfam is also often used as a source of individual ncRNA sequences.
As described in previous Rfam publications, the database is built upon well-curated seed alignments of representative members of an RNA family (6–8). These are used to build CMs, statistical models of a family's conserved sequence and secondary structure, using the Infernal suite of analysis tools (9). The resultant covariance models are used to scan a large database of nucleotide sequences that is derived from the EMBL nucleotide archive (10). The searches return a list of putative homologs, or hits, ranked by bit-scores derived from the CMs. A hit's bit-score is the log odds ratio of the probability the hit was generated by the CM versus a random model of background sequence. An expert curator provides a threshold that in their opinion best discriminates between bona fide homologs to the seed sequences and the background distribution of false hits. Subsequently, all sequences with a bit-score above the threshold are included in an automatically generated alignment to the CM.
NEW DEVELOPMENTS
The Rfam 10.0 “decimal” release
In order to keep Rfam as up-to-date as possible we aim to make regular releases of the database. These releases are snap-shots of the live, internal version of the database that are made publicly available via the websites and ftp. We have two types of release. A major release (indicated by an integer and a ‘.0’ in the version number e.g. ‘10.0’) usually involves updating the underlying sequence database, Rfamseq, to the latest version of EMBL and remapping all the seed sequences to the new databases. All the families are subsequently searched against the new database and, if necessary, re-thresholded. Minor releases are indicated by ‘.1’, ‘.2’, etc. in the version number e.g. ‘10.1’. These are usually made after adding many new families to the database built on the same underlying sequence database.
Rfam 10.0 was released in early 2010. This release included a major update to the underlying search algorithm, switching to a new version of Infernal, v1.0 (9). This required individually re-thresholding each Rfam family due to an important change in Infernal’s underlying scoring scheme from maximum likelihood alignment scores to summed scores over all possible alignments [i.e. switching from using the CYK algorithm to the Inside algorithm (11)]. Additionally, the new version of Infernal reports estimates of the statistical significance of hits (E-values) returned from database searches using Rfam 10.0 CM files. We also mapped all the families and searched a new version of Rfamseq based on EMBL 100 (10). The result of these and other internal improvements to our pipeline resulted in a 178% increase in the number of regions that Rfam covers, which contrasts with the rather modest increase in the size of Rfamseq by 40%. This has caused some of our alignments to become very large. For example, the tRNA full alignment now contains more than 1 million sequences. The amount of compute required for this release was roughly 5 CPU months to calibrate the models, 1 CPU year to run blast, 3 CPU years to run CM-searches (cmsearch) and 15 CPU days to produce CM-derived multiple sequence alignments (cmalign).
Evaluating the success of the Wikipedia community annotation model
One of the fundamental problems facing any biocuration effort is keeping the annotation of the entities stored in a database up to date with the current literature. Typically, the annotation of existing entries changes less quickly than new data are added, so entries become rapidly out-of-date.
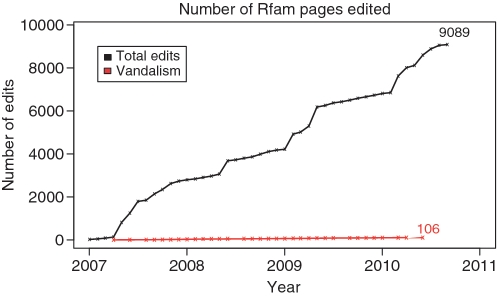
In mid-2007, Rfam began experimenting with using Wikipedia as a means for storing and curating the textual annotation of RNA families. Three years on, the RNA family pages have received more than 9000 edits from more than 1000 unique users. Slightly over 1% of these edits have been recognized as possible vandalism (Figure 1). The resulting marked-up annotation and curated references has dramatically improved the content of the Rfam database compared with the pre-2007 static text. The Wikipedia entries also help drive users to the Rfam website. Approximately 15% of all the web-traffic to http://rfam.sanger.ac.uk now comes via Wikipedia. As has been observed by others, a typical Google search for a biological term returns a Wikipedia entry among the top hits (12,13). From a curator’s viewpoint, Wikipedia is an excellent model to take advantage of as it includes a large community of contributors and comes with a number of user-friendly tools that help with basic editing, maintaining references and automated updates to pages with programs called bots. The large community also has other benefits, such as the well documented long-tail effect, where the majority of new content is added by a large number of editors, each of whom makes just a few edits (12,13). There are also dedicated editors who are obsessed with small but important details that an average curator may not have time to attend to, such as consistency of style, grammar and spelling. There are also editors who are dedicated to reverting obvious non-constructive edits, commonly referred to as `vandalism’, which are usually recognized and reverted within seconds. It is important to note that all edits are reviewed before appearing on the Rfam website, so the amount of overt vandalism reaching Rfam is 0. Given our positive experiences, we can highly recommend other curation efforts turning to Wikipedia for their annotation. However, it must be borne in mind that Wikipedia is built by consensus and to gain its benefits you will lose the tight control of the data allowed by in-house curation.
Figure 1.
Edits for Wikipedia articles on RNA families. The cumulative number of edits since 1st January 2007 for the 733 Wikipedia articles that are associated with Rfam entries is shown in black. The total number of edits that were reverted or labeled as vandalism is shown in red. To mid-2010, there were just 106 of these. However, some reverted edits may have been well-intentioned but were deemed inappropriate for Wikipedia.
Rfam clans
One of the fundamental quality control steps that Rfam employs is that no two families can annotate the same nucleotide. This rule prevents us building two or more families for essentially the same entity. When building new Rfam families or extending an existing family, we sometimes find ourselves artificially increasing the threshold to avoid overlaps with another family or trimming the ends of families that have incorrect boundaries. We also find that a single alignment may not capture all the diversity of a group of homologous RNAs. To resolve some of these issues, we have borrowed the concept of a clan from the MEROPS and Pfam databases (14,15).
We have added 99 clans for the Rfam 10.0 release. These clans describe explicit relationships between families that either clearly share a common ancestor but are too divergent to be reasonably aligned or groups of families that could be aligned, but have clearly distinct functions and therefore should be kept as separate families. For example, the RNase P clan contains five homologous families RNase MRP, archeal RNase P, nuclear RNase P and the bacterial RNase P, types a and b. These RNAs are ribozymes involved in processing of pre-tRNA and pre-rRNA sequences. The RNase Ps are, however, notoriously difficult to align to each other. Furthermore, RNase P and RNase MRP are functionally distinct molecules (16). Another clan of interest is Glm; this clan contains two homologous but functionally distinct bacterial small RNAs, GlmY and GlmZ, which act in a hierarchical fashion to regulate the translation of the glmS coding gene. GlmY activates expression of GlmZ which in turn de-sequesters the GlmS Shine-Dalgarno sequence via an anti-antisense interaction (17). The new clans mean that some of the internal quality control measures that Rfam uses can be relaxed for the clanned families. Primarily this means we can ignore our no-overlap rule, which has meant that in the past some of these families have had artificially high thresholds to avoid overlapping a related but distinct family.
In order to help assess the likelihood of a relationship between two or more families, we used a number of independent lines of evidence. These included sequence analysis based upon a SCOOP-like analysis for comparing overlapping hits from both profile hidden Markov model (HMM) and covariance model searches (18), the profile-profile comparison tool PRC (19) and literature searches for functional and evolutionary relationships. For the snoRNA and miRNA families, we were able to utilize some additional sources of information in order to establish homology. For the snoRNAs, we used some of the specialized snoRNA databases to confirm whether families targeted orthologous regions of rRNA, for many snoRNAs this helped to confirm a relationship between the families (20–23). For the miRNAs, we used the annotated seed region of the mature miRNA (24). If two or more miRNA families shared a significant amount of similarity in the seed region, and if they had further similarities identified by the sequence analysis tools, then these too were added to clans.
Species labels
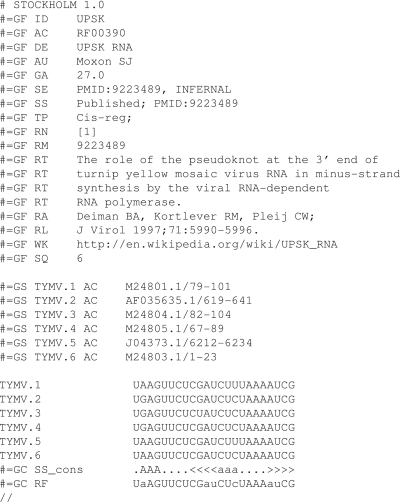
The new set of seed and full alignments available via the website use descriptive species labels for sequence names rather than the more cryptic EMBL accessions and coordinates that were previously provided. The provenance of the sequence data is maintained by using ‘#=GS’ tags from Stockholm format (25) to provide a mapping back to EMBL accessions (Figure 2). Stockholm is a versatile markup format for biological sequence alignments. It allows the markup of general file information, including references, comments and cross-links. It also allows the mark-up of regions of an alignment that cannot be aligned with tildes in the ‘#=GC RF’ lines.
Figure 2.
An example Stockholm alignment for the UPSK pseudoknot from turnip yellow mosaic virus. The Stockholm alignment format is flexible enough to allow generic mark-up of file information with ‘#=GF' lines, sequence information with ‘#=GS' lines and column information with ‘#=GC' lines. Each is followed by at least a two-letter code giving an indication for what follows e.g. ‘ID' implies ‘identifier', ‘AC' implies ‘accession', ‘AU' implies ‘author', etc. All the commonly used tags are documented in the Wikipedia article for Stockholm alignment (25).
Ontologies
An important feature for any biocuration effort is linking to related resources, for example, primary sequence resources databases, genomes and to specialized resources such as miRBase and the snoRNA databases. Recently, a number of groups have started developing controlled vocabularies for describing biological entities. Two efforts of particular relevance to Rfam are the sequence ontology (SO) and the gene ontology (GO) (26,27). For the majority of Rfam families, we have now added cross-links to both the SO and the GO. Many of these were provided by researchers at the functional RNA database (28). In the near future, we plan to introduce more ncRNA terms back into the ontologies. Until then the mapping will remain rather coarse-grained and closely related to the existing types Rfam uses as annotation (6). This mapping groups the RNAs into three main groups: ‘cis-reg’, ‘gene’ and ‘intron’ with subtypes such as ‘riboswitch’, ‘miRNA’ and ‘snoRNA’.
Future developments
New families in Rfam 10.1
For the forthcoming minor release of Rfam, we have added a number of new and notable families. Of particular note are the direct submissions of Stockholm formatted alignments and corresponding Wikipedia articles from the RNA community via the RNA families track at RNA Biology (8). This track has released much of the burden of building these new families from our curators, and the families produced have been built and annotated by experts and are therefore of high quality. Updated families from this route include RNase MRP, SRP, tmRNA and the U3 snoRNA (29–32). In addition, several families missing from past Rfam releases have been published, including the SmY RNA, the cyanobacterial RNA Yfr2, several Trypanosomatid snoRNAs, the self-splicing ribozyme GIR1, an influenza pseudoknot, the Staphylococcus small RNA RsaOG and a putative RNA antitoxin, ptaRNA1 (33–39). The ptaRNA1 article alerted us to the fact that Rfam contains none of the published and well-characterized RNA antitoxins such as sok and symE (40). These omissions will be remedied in Rfam 10.1. A growing class of cis-regulatory elements are the environmental sensors. These are generally structured 5′ UTR elements that change conformation in response to environmental changes such as temperature or pH; this change subsequently influences the expression of the protein encoded in the host mRNA. We have added the first examples of a cold sensor and a pH sensor (41,42). Finally, we have received a dramatic number of submissions from a recent bioinformatic screen that was followed by a thorough analysis of the predictions largely based upon genomic context. This has resulted in more than 80 new additions to the database (43). Fortunately, the authors kindly provide both Stockholm formatted alignments and Wikipedia articles for these new families.
Covariance model pre-filters
A pressing issue for Rfam is the replacement of WU-BLAST as a pre-filter for searching the Rfamseq database. The legal rights to up-to-date versions of WU-BLAST were recently acquired by a commercial entity and the software can no longer be considered free in any meaningful sense. However, there have been several developments that should allow profile HMMs to be used as effective pre-filters for covariance model searches (44). Accelerated profile HMM searches are now available through the HMMER package (45–47). In the near future, Rfam will therefore be in a position to replace the current BLAST-based filters with accelerated profile HMMs.
Scale
Sequencing projects such as the Genome 10K (48) and other attempts to fill sequencing gaps in the tree of life (49) mean that most Rfam families will dramatically increase in depth in the near future. Large alignments already pose a considerable challenge when it comes to displaying or distributing the alignments themselves, or building and displaying related data such as species and phylogenetic trees. Novel techniques will need to be developed in order to deal with these and many other issues of scale. We look forward to working with the wider community to develop these new tools and techniques.
FUNDING
Wellcome Trust (grant number WT077044/Z/05/Z) (to P.P.G., J.D., J.T., I.H.O., B.M. and A.B.); Howard Hughes Medical Institute (R.D.F, E.P.N., D.L.K. and S.R.E); University of Manchester (S.G.J.). Funding for open access charge: The Wellcome Trust (grant number WT077044/Z/05/Z).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Many thanks to Guy Coates, James Beal and Peter Clapham for assistance with improving the performance of computational and software infrastructure. The authors received invaluable feedback at the 2009 Benasque RNA Workshop.
REFERENCES
- 1.Holmes I. A probabilistic model for the evolution of RNA structure. BMC Bioinformatics. 2004;5:166. doi: 10.1186/1471-2105-5-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Do CB, Woods DA, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:e90–e98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 3.Yao Z, Weinberg Z, Ruzzo WL. CMfinder–a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Buhler J. Designing secondary structure profiles for fast ncRNA identification. Comput. Syst. Bioinformatics Conf. 2008;7:145–156. [PubMed] [Google Scholar]
- 5.Yusuf D, Marz M, Stadler PF, Hofacker IL. Bcheck: a wrapper tool for detecting RNase P RNA genes. BMC Genomics. 2010;11:432. doi: 10.1186/1471-2164-11-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33(Database issue):D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37(Database issue):D136–D1340. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leinonen R, Akhtar R, Birney E, Bonfield J, Bower L, Corbett M, Cheng Y, Demiralp F, Faruque N, Goodgame N, et al. Improvements to services at the European Nucleotide Archive. Nucleic Acids Res. 2010;38(Database issue):D39–D45. doi: 10.1093/nar/gkp998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eddy SR. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics. 2002;3:18. doi: 10.1186/1471-2105-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huss JW, Orozco C, Goodale J, Wu C, Batalov S, Vickers TJ, Valafar F, Su AI. A gene wiki for community annotation of gene function. PLoS Biol. 2008;6:e175. doi: 10.1371/journal.pbio.0060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huss JW, Lindenbaum P, Martone M, Roberts D, Pizarro A, Valafar F, Hogenesch JB, Su AI. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2010;38(Database issue):D633–D639. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem. J. 1993;290(Pt 1):205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34(Database issue):D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis JC, Brown JW. The RNase P family. RNA Biol. 2009;6:362–369. doi: 10.4161/rna.6.4.9241. [DOI] [PubMed] [Google Scholar]
- 17.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bateman A, Finn RD. SCOOP: a simple method for identification of novel protein superfamily relationships. Bioinformatics. 2007;23:809–814. doi: 10.1093/bioinformatics/btm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madera M. Profile Comparer: a program for scoring and aligning profile hidden Markov models. Bioinformatics. 2008;24:2630–2631. doi: 10.1093/bioinformatics/btn504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samarsky DA, Fournier MJ. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:161–164. doi: 10.1093/nar/27.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JW, Echeverria M, Qu LH, Lowe TM, Bachellerie JP, Hüttenhofer A, Kastenmayer JP, Green PJ, Shaw P, Marshall DF. Plant snoRNA database. Nucleic Acids Res. 2003;31:432–435. doi: 10.1093/nar/gkg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li SG, Zhou H, Luo YP, Zhang P, Qu LH. Identification and functional analysis of 20 Box H/ACA small nucleolar RNAs (snoRNAs) from Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:16446–16455. doi: 10.1074/jbc.M500326200. [DOI] [PubMed] [Google Scholar]
- 23.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34(Database issue):D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stockholm format. http://en.wikipedia.org/wiki/Stockholm_format Stockholm format (19 June 2010, date last accessed)
- 26.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37(Database issue):D89–D92. doi: 10.1093/nar/gkn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dávila López M, Rosenblad MA, Samuelsson T. Conserved and variable domains of RNase MRP RNA. RNA Biol. 2009;6:208–220. doi: 10.4161/rna.6.3.8584. [DOI] [PubMed] [Google Scholar]
- 30.Rosenblad MA, Larsen N, Samuelsson T, Zwieb C. Kinship in the SRP RNA family. RNA Biol. 2009;6:508–516. doi: 10.4161/rna.6.5.9753. [DOI] [PubMed] [Google Scholar]
- 31.Mao C, Bhardwaj K, Sharkady SM, Fish RI, Driscoll T, Wower J, Zwieb C, Sobral BW, Williams KP. Variations on the tmRNA gene. RNA Biol. 2009;6:355–361. doi: 10.4161/rna.6.4.9172. [DOI] [PubMed] [Google Scholar]
- 32.Marz M, Stadler PF. Comparative analysis of eukaryotic U3 snoRNA. RNA Biol. 2009;6:503–507. doi: 10.4161/rna.6.5.9607. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Otto W, Marz M, Eddy SR, Stadler PF. A survey of nematode SmY RNAs. RNA Biol. 2009;6:5–8. doi: 10.4161/rna.6.1.7634. [DOI] [PubMed] [Google Scholar]
- 34.Gierga G, Voss B, Hess WR. The Yfr2 ncRNA family, a group of abundant RNA molecules widely conserved in cyanobacteria. RNA Biol. 2009;6:222–227. doi: 10.4161/rna.6.3.8921. [DOI] [PubMed] [Google Scholar]
- 35.Doniger T, Michaeli S, Unger R. Families of H/ACA ncRNA molecules in trypanosomatids. RNA Biol. 2009;6:370–374. doi: 10.4161/rna.6.4.9270. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen H, Johansen SD. Group I introns: moving in new directions. RNA Biol. 2009;6:375–383. doi: 10.4161/rna.6.4.9334. [DOI] [PubMed] [Google Scholar]
- 37.Gultyaev AP, Olsthoorn RC. A family of non-classical pseudoknots in influenza A and B viruses. RNA Biol. 2010;7:125–129. doi: 10.4161/rna.7.2.11287. [DOI] [PubMed] [Google Scholar]
- 38.Marchais A, Bohn C, Bouloc P, Gautheret D. RsaOG, a new staphylococcal family of highly transcribed non-coding RNA. RNA Biol. 2010;7:116–119. doi: 10.4161/rna.7.2.10925. [DOI] [PubMed] [Google Scholar]
- 39.Findeiss S, Schmidtke C, Stadler PF, Bonas U. A novel family of plasmid-transferred anti-sense ncRNAs. RNA Biol. 2010;7:120–124. doi: 10.4161/rna.7.2.11184. [DOI] [PubMed] [Google Scholar]
- 40.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, Gualerzi CO, Pon CL. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell. 2010;37:21–33. doi: 10.1016/j.molcel.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 42.Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome. Biol. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberg Z, Ruzzo WL. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics. 2006;22:35–39. doi: 10.1093/bioinformatics/bti743. [DOI] [PubMed] [Google Scholar]
- 45.Eddy SR. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 2008;4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 47.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genome 10K Community of Scientists, C. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J. Hered. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, et al. The RNA structure alignment ontology. RNA. 2009;15:1623–1631. doi: 10.1261/rna.1601409. [DOI] [PMC free article] [PubMed] [Google Scholar]