Abstract
The SUPERFAMILY resource provides protein domain assignments at the structural classification of protein (SCOP) superfamily level for over 1400 completely sequenced genomes, over 120 metagenomes and other gene collections such as UniProt. All models and assignments are available to browse and download at http://supfam.org. A new hidden Markov model library based on SCOP 1.75 has been created and a previously ignored class of SCOP, coiled coils, is now included. Our scoring component now uses HMMER3, which is in orders of magnitude faster and produces superior results. A cloud-based pipeline was implemented and is publicly available at Amazon web services elastic computer cloud. The SUPERFAMILY reference tree of life has been improved allowing the user to highlight a chosen superfamily, family or domain architecture on the tree of life. The most significant advance in SUPERFAMILY is that now it contains a domain-based gene ontology (GO) at the superfamily and family levels. A new methodology was developed to ensure a high quality GO annotation. The new methodology is general purpose and has been used to produce domain-based phenotypic ontologies in addition to GO.
INTRODUCTION
SUPERFAMILY (1) is a publicly available resource that provides the prediction of protein domains of known structure in amino acid sequences. The database contains a periodically updated library of expert-curated hidden Markov models (HMM) representing all protein domains of known structure. The classification of these domains is taken from the structural classification of protein (SCOP) database (2). SCOP groups protein domains hierarchically, according to their nature of similarity (sequence, evolutionary and structural), into Class, fold, superfamily and family. The SUPERFAMILY database is particularly focused on the superfamily level. Two domains are put in the same superfamily (or evolutionary) level if, and only if, there is structural functional and sequence evidence for a common ancestor (3).
The SUPERFAMILY website (http://supfam.org) offers a variety of methods to analyze proteins and superfamilies. A keyword search facility is available from all pages on the website. At the genomic level the user can investigate under- and over-represented superfamilies (3), phylogenetic trees, domain architectures and networks (4) and examine the distribution of superfamilies across the tree of life (5).
Here, we describe several improvements introduced into the SUPERFAMILY 1.75 release of the database since last publication (5). In the next section we summarize the updates in SUPERFAMILY 1.75, then we describe new features incorporated in the database as well as improvements in the back end and underlying procedure. In the last section we explain in detail a new procedure to create domain-centric functional and phenotypic annotations from individual protein-level annotations.
SUMMARY OF THE UPDATES
The most significant advance is that SUPERFAMILY now contains domain-based gene ontology annotation (GOA) at both the family and superfamily levels. To obtain a high-quality GOA with associated significance scores it was necessary to develop a novel methodology. The new methodology is of general use and we have already further applied it ourselves to provide SUPERFAMILY with domain-based phenotypic ontology in addition to GO.
On the protein structure side we have not only extended the HMM library to be up to date with the current release of SCOP (1.75), but a major addition is the entire class of ‘coiled coil’ proteins which was until now excluded. On the sequence side, we remain up to date with the rapidly growing number of genomes and the expanding size of UniProt (6); we have expanded to include over 120 meta-genomes from environmental sequencing projects, and have explicitly added 2354 plasmids and over 2473 viral genomes and their taxonomy.
The SUPERFAMILY reference tree of (sequenced) life has been significantly improved: it now uses a probabilistic method constrained by known taxonomy with organisms ordered intuitively. The visualization tool now has the facility to highlight a chosen superfamily, family or domain architecture on the tree of life. In fact in combination with the GO it is now possible to annotate the tree of life with any chosen function term.
In addition to the outwardly facing changes listed above, there have been major changes to the internal structure of the resource that will affect users. Our (as yet unpublished) analysis has shown that the HMM scoring component of the new HMMER3 (7) produces superior results to that of SAM (8) and therefore, we have converted to HMMER3 scoring. HMMER3 is orders of magnitude faster than SAM or HMMER2 and has enabled us to move to a software cloud-based pipeline. In fact we make available an Amazon web services (AWS) elastic computer cloud (EC2) instance image eliminating the need for users of the package to install or run locally. We have also re-designed the fundamental internal SQL database structure so that sequences and their assignments have only one instance; however, many genomes or sequence sets they participate in.
The tree of (sequenced) life
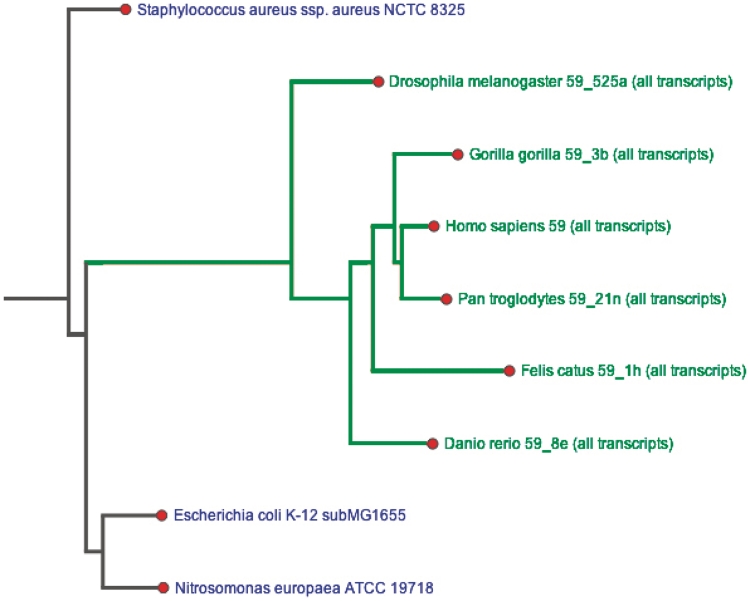
The SUPERFAMILY reference species tree of (sequenced) life is now generated with RAxML (9) using the gamma model of rate heterogeneity. Trees are constrained to the NCBI taxonomy (10) and plotted online as scalable vector graphics (SVG) using an extended version of TreeVector (11), and can be downloaded in a variety of formats. Superfamilies, families, architectures, GO terms and phenotype terms can now be highlighted individually or as combinations on the phylogenetic tree of all sequenced genomes, visually illustrating the evolutionary history of the set across the tree of sequenced life (Figure 1). Users can choose to highlight over the entire tree of all sequenced genomes, the highest tree node containing all highlighted species, or a manually selected list containing solely the clade of interest. In addition, TreeVector can now assign priorities to leaf nodes so that species/clades of greater interest appear at the top of the tree. Trees are linked to at the bottom of the ‘Taxonomic Distribution’ tab and from the GO and phenotype terms pages.
Figure 1.
Presence/absence of the fibronectin type III superfamily in selected genomes by automatic highlighting of branches of the phylogenetic tree that contain the superfamily in green.
Text query searches
In the SUPERFAMILY 1.75 release we added a full text query for SCOP descriptions and species names resulting in more efficient and accurately ranked search results. The relevance score returned by full text queries is used to rank the search results.
The search functionality covers SCOP descriptions, species names including common names plus sequence, SUPERFAMILY HMM, SCOP, PDB (12) and InterPro (13) identifiers. For multi-word searches an `entire phrase’ (aka AND) search is performed first. An `any word’ (aka OR) search is perfomed if the entire phrase search fails. Common words like `and’, `the’ are removed. Commonly occurring but specialized words like `domain’, `superfamily’, `protein’ and `gene’ will also be removed.
Coiled coils
HMMs for the domains in the previously unused SCOP coiled-coil class have been included in the SUPERFAMILY HMM library facilitating the production of a sister resource, Spiricoil (http://supfam.org/SUPERFAMILY/spiricoil/) that deals with the evolution and identification of this super-secondary structure.
It has long been believed that coiled coils are a problem class due to their tendency to contain low complexity or repeat regions. Because of this they have previously been omitted in terms of homology-based domain prediction. We have, however, succeeded in constructing new HMMs covering all of the domains belonging to the 55 superfamilies in this class. These have now been added to the SUPERFAMILY HMM library and are integrated into the SUPERFAMILY pipeline.
We also identified all coiled coil containing families and superfamilies in other classes. Annotation of the position and oligomeric state of the coiled coils from those structures and from the SCOP coiled class were used within Spiricoil (14) to explore their evolution across genomes as well as to enable prediction of their oligomeric state.
Cloud computing
Computer performance improvements generally follow ‘Moore’s Law’; doubling every 18–24 months. Sequencing throughput, on the other hand, has a 5-fold growth rate per year (15). With new sequencing technologies such as the third generation sequencing we will soon be able to scan entire genomes, microbiomes and transcriptomes and assess epigenetic changes directly in just minutes and for an affordable price (16,17). Large-scale projects such as the 1000 genomes project are already generating petabytes of raw information (18).
Due to the ever-growing number of genomes we implemented our pipeline for assigning protein domains, from all completely sequenced genomes, in the Amazon EC2 cloud (see Tables 1 and 2 for details). The cloud not only allows us to analyze the genomes more quickly but also provide a scalable source of computing power to guarantee the future provision of the SUPERFAMILY resource to the community. We provide a publicly available cloud image with our pipeline, allowing users to run the assignment analysis on their own genomes very simply using the Amazon EC2 cloud. The image is provided automatically via E-mail upon registration for the SUPERFAMILY package and downloads.
Table 1.
SUPERFAMILY 1.75
| Release date | September 2010 |
|---|---|
| Number of HMM models | 15 438 |
| Number of completely sequenced genomes, strains and collections | 1628 |
| Eukaryotes | 341 |
| Archeabacterial | 87 |
| Eubacterial | 1077 |
| Metagenomes | 118 |
| Plasmids | 2354 |
Table 2.
SUPERFAMILY 1.75 statistics
| Protein with assignments (%) | Amino acid coverage (%) | |
|---|---|---|
| Eukaryotes | 59.11 | 38.9 |
| Archeabacteria | 65.13 | 61.67 |
| Eubacteria | 68.08 | 63.4 |
| Uniprot | 64 | 56 |
| Metagenomes | 51.47 | 54.1 |
| Plasmids | 47 | 47 |
HMMER3
The HMMER software package for hidden Markov model (HMM) analysis of biological sequences recently underwent a major new release. The scoring component of the new HMMER3 package performs better than either HMMER2 or SAM when tested against the SCOP database (as yet unpublished); the two older packages were previously shown by us to be of similar performance when it comes to scoring (19). Amazingly, HMMER3 scoring is also orders of magnitude faster. For these reasons, and because HMMER is more commonly used by others, we have converted our pipeline and web services to use HMMER3 scoring. This should be of great benefit to users of the HMM library and is a significant contribution to the sustainability of the resource. To complement the increased speed of HMM scoring we have also streamlined and accelerated the post-processing software and added multi-threading support. The limit for submitting to the web server for processing has consequently been increased from 20 sequences to 1000 sequences.
New structures and sequences
Since the last update of SUPERFAMILY was published the database has moved to the current 1.75 version of SCOP. This includes the addition of nearly 200 new superfamilies and nearly 500 families represented by 1392 new HMMs. All of the genome assignments have been recalculated on the new model library.
New genomes are continually added to SUPERFAMILY as they become available. We also now provide assignments to all structures not yet in SCOP, automatically updated weekly with the protein data bank (PDB). Since the last publication we have added over 400 genomes, 120 metagenomics sequence sets and we now explicitly list over 2473 viruses and 2355 plasmids. There are now ∼30 million sequences in the database (including redundancy with UniProt). The percentage of sequences with an assignment in a genome has increased slightly, but the total amino acid sequence coverage has in some cases dropped slightly. The latter is due to a characteristic of HMMER3. The number of domains assigned in each genome has increased, but the average length has decreased (Table 2).
DOMAIN-CENTRIC FUNCTIONAL AND PHENOTYPIC ANNOTATIONS
A full understanding of a protein’s functions requires knowledge of its building blocks, particularly functional aspects of 3D structural domains. This knowledge is also vital to make sense of the sequenced genomes and their evolution. The promising use in comparative and functional genomics, of domain-centric functional annotations lags far behind the protein-level annotations. By convention, functional annotations are assigned onto individual proteins ignoring the context of the structural domains. For instance, the GOA project (20) provides high-quality GOAs directly associated to proteins in the UniProt Knowledgebase (UniProtKB) (6) over a wide spectrum of species. Even worse, the lack of comprehensive structural domain information further discourages functional annotations at the domain level, although the number of experimentally resolved structural proteins deposited in PDB (21) continues to increase. Fortunately, the domain annotations of a protein can be routinely assigned using HMMs (22,23) based on SCOP (24). For example, the current SUPERFAMILY database provides high-coverage domain assignments for proteins in UniProtKB. Beyond that, we have recently taken advantage of manually derived GOA and comprehensive domain assignments for proteins in UniProtKB, to statistically infer domain-centric functional annotations. Such statistical inference is based on the assumption: if a GO term tends to annotate proteins containing a domain, then such a term should also confer functional signals for that domain. Respecting the hierarchical structure of GO as well as the domain composition of proteins, we have generated the first GOAs for evolutionarily close domains (at the SCOP family level) and distant domains (at the SCOP superfamily level). Here we emphasize the GOAs of domains at SCOP family and superfamily levels, which will greatly enrich the other existing resources (13,25,26). In particular, we expect that the domain mappings between SCOP and InterPro (and Pfam) will benefit each other in terms of their relevance to GO. Moreover, we have initialized a trimmed-down version of GO, which is the most informative for annotating domains. This resource represents an ongoing effort to develop a structural domain functional ontology (SDFO). We expect domain-centric GOAs, together with other resources and tools in the SUPERFAMILY web server, will greatly facilitate our understanding of functional genomics across the tree of life.
The strategy described above can easily be generalized for detecting other ontology relatedness to structural domains. For example, structural domains can bridge gaps between sequences of proteins and their phenotypic outcomes. We reason that proteins sharing the same structural domain, upon being genetically disrupted, lead to certain phenotypes probably related to a mutation occurring in that domain. Based on this, domain-centric phenotypic annotations can be similarly inferred from the mammalian phenotype ontology (MPO) curated by the mouse genome informatics (MGI) (27) and from the Human Phenotype Ontology (HPO) which is built on the online Mendelian inheritance in man (OMIM) (28). Promisingly, domain-centric phenotypic annotations can serve as an alternative starting point to explore genotype–phenotype relationships.
Functional and phenotypic annotations of structural domains at the SCOP superfamily and family levels
The GO and phenotype data are available for download and accessible via the web interface by selecting from the navigation bar on the left or in the ‘structural classification’ tab on the page for an individual superfamily or family.
The pipeline of building domain-centric GOAs
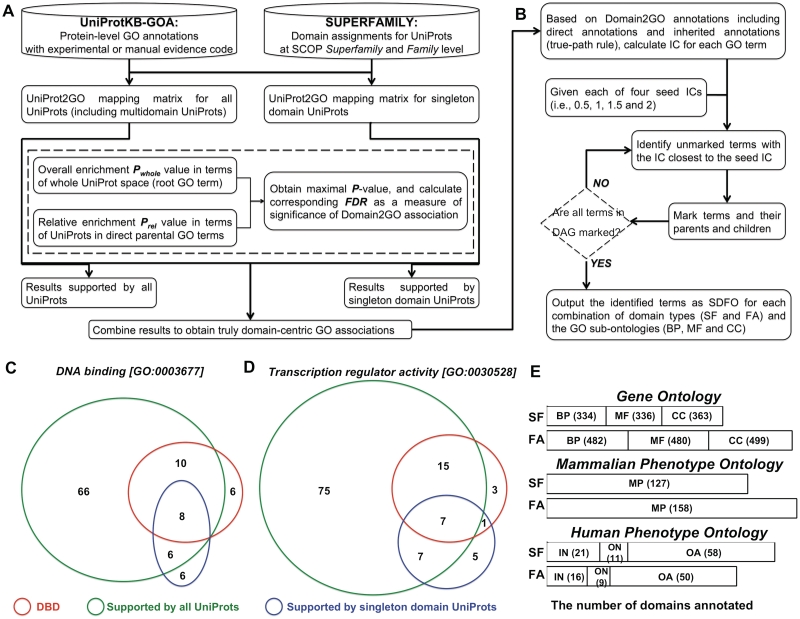
The procedures to create domain-centric GOAs from individual protein-level annotations are summarized in Figure 2. The motivations behind them are: (i) from the biological point of view, structural domains constitute functional units of proteins and thus their functions are inherent in the protein-level GOAs; (ii) from the methodological point of view, such inherent GOAs for a domain can be reversely inferred if the number of domain-containing GO-annotated proteins is significantly higher than would be expected by chance. To realize the motivations, we started with the primary sources of high-quality GOAs for individual proteins as well as their high-coverage domain compositions (as assigned by HMMs in SUPERFAMILY) at SCOP family and superfamily levels. Following tests based on the hypergeometric distribution, two types of enrichments were performed to infer the overall and relative associations between a domain and a GO term; two sets of proteins (i.e. singleton domain proteins and all proteins including multiproteins) were used to support associations. To make sure that each association is truly domain-centric, we combined the results from the tests above. To the best of our knowledge, these GOAs are the first resource tailored to evolutionarily close domains (at the SCOP family level) and distant domains (at the SCOP superfamily level), which can be further used to initialize a SDFO.
Figure 2.
Functional and phenotypic annotations of structural domains at the SCOP superfamily (SF) and family (FA) levels. (A) Flowchart of inferring domain-centric GOAs using UniprotKB-GOA database and domain assignments in SUPERFAMILY database. (B) Illustration of the procedure to create SDFO based on information theoretic analysis of Domain2 GOA profiles. (C) Venn diagram in which the area of each region is proportional to the differences and intersections among domains annotated to a GO term `DNA binding’ [GO:0003677] using all UniProt sequences (90, circled in green), domains annotated to the term only using singleton domain UniProt sequences (20, circled in blue), and domains in DBD which can be found in at least one UniProt sequence annotated to the term (24, circled in red). (D) Venn diagram showing the differences and intersections among domains annotated to a GO term `transcription regulator activity’ [GO:0030528] using all UniProt sequences, only using singleton domain UniProt sequences, and in DBD which can be found in at least one UniProt sequence annotated to the term. (E) The total number (shown in parenthood) of domains annotated to ontologies. GO depicts three biological concepts: BP, Biological Process; MF, Molecular Function; CC, Cellular Component. Results are based on Domain2 GOAs supported both by singleton domain UniProt sequences and all UniProt sequences. In MPO, it describes mammalian phenotype (MP) related to the mouse with a specific genetic mutation. HPO has three sub-ontologies: IN, inheritance; ON, onset and clinical course; OA, organ abnormality.
GOAs of proteins in UniProtKB and their domain assignments by SUPERFAMILY
The primary source of protein-level GOAs is taken from UniProtKB-GOA. To reduce false-positives and avoid data circularity from InterPro (13) and Pfam (29), we only consider those annotations supported by experimental or manual evidence codes (30). Due to the availability of relatively complete domain assignments for UniProt provided by SUPERFAMILY, filtering GOA does not shrink our analyzable UniProt sequence space (i.e. those UniProt sequences annotated with at least one GO term and containing at least one domain). Notably, the large UniProt sequence space in this study allows us to ensure that statistical inference has adequate power to reveal significant associations between a GO term and a domain from protein-orientated GOAs.
Inferring domain-centric GOAs
Given sets of UniProt sequences with manually derived GO terms from GOA as well as their structural domains assigned by SUPERFAMILY, potential associations between a GO term and a domain can be reversely inferred by examining whether the observed number of domain-containing GO-annotated UniProt sequences is significantly higher than would be expected by chance. The statistical significance of inference is assessed based on the hypergeometric distribution, followed by multiple hypotheses testing in terms of false discovery rate (FDR). More importantly, we have addressed two issues regarding the hierarchical structure of GO and the nature of domain composition for multi-domain proteins.
First, we respect the hierarchical structure of GO, which is organized as a directed acyclic graph (DAG) by viewing individual terms as a node and its relations to parental terms (allowing for multiple parents) as directed edges. Specifically, we perform statistical inference of possible associations between a GO term (say t) and a domain (say d), not only in terms of all UniProt sequences [see Equation (1)], but also in the context of those UniProt sequences annotated to all direct parents of that GO term [see Equation (2)], These dual constraints [see Equation (3)], ensure that only those most informative GO terms are retained. When simultaneously comparing multiple hypothesis tests, statistical significance of domain-GO term associations can be assessed by the method of FDR (31) [see Equation (4)]. The resultant FDR was used to determine the significance of domain-GO term associations.
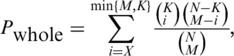 |
(1) |
where N is the number of UniProt sequences annotated with at least a GO term and containing at least a domain, M for the number of UniProt sequences containing the domain d, K for the number of UniProt sequences annotated with the GO term t, X for the observed number of UniProt sequences annotated with the GO term t as well as containing the domain d, and Pwhole is the expected probability of observing X or more UniProt sequences under the hypergeometric distribution.
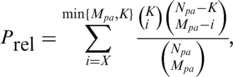 |
(2) |
where Npa is the number of UniProt sequences annotated with all direct parents of that GO term t in DAG, Mpa for the number of UniProt sequences containing the domain d after intersecting with those UniProt sequences in Npa, K for the number of UniProt sequences annotated with the GO term t, X for the observed number of UniProt sequences annotated with the GO term t as well as containing the domain d, and Prel is the expected probability of observing X or more UniProt sequences under the hypergeometric distribution.
| (3) |
where P is defined as the maximum P-values in terms of overall enrichment test and relative enrichment test.
| (4) |
where FDR is calculated using the Benjamini–Hochberg (BH) derived step-up procedure, Pir is the i-th ranked P in an ascending manner, L for the number of all possible domain-GO term associations.
Second, we respect the nature of the domain composition of proteins. The contribution of each domain in a multi-domain protein to its functions may be dominant or trivial or between. Because we here aim to generate truly domain-centric functional annotations, the resulting GO terms for a given domain should account for both singleton domain proteins and multi-domain proteins containing that domain. To such end, we calculated significance [FDR, see Equations (1–4)] of associations only using singleton domain UniProt sequences and using all UniProt sequnces (including multi-domain sequences). The criteria for identifying the domain-GO associations were based on stringent FDR (<0.001), supported both by singleton domain sequences and all sequences.
Since GO depicts three complementary biological concepts including biological process (BP), molecular function (MF) and Cellular Component (CC), and SCOP classifies evolutionary-related domains into superfamily level and family level, we have accordingly generated the domain-centric GOAs for each of the three concepts at the two domain levels.
Initializing SDFO
Here, we are aiming to get lists of GO terms that are the most informative in terms of annotating structural domains. To do so, we applied information theory to define information content (IC) of a GO term based on domain-GOA profiles [see Equation (5)]. For any domain, GO terms annotated to that domain constitute a domain-GOA profile in DAG, including direct annotations as well as inherited annotations according to the true-path rule. Considering the nature of dependencies among GO terms (or so-called true-path rule), a domain/protein directly annotated to a specific GO term (termed as direct annotations) should be inheritably annotated to its parental terms (terms as inherited annotations). GOAs generated above can be considered as direct annotations. The complete GOAs (direct and inherited) are used to calculate IC for all GO terms. Actually, the IC of a GO term gives a measure of how informative it is to annotate all annotatable domains. For example, a GO term (i.e. BP or MF or CC) with an IC of 0 would be expected to annotate all domains. More importantly, those GO terms with similar IC can represent a partition that has been used to produce the GO partition database (32). Similarly, we have developed a procedure (Figure 2B) to create meta-GO terms as a proxy for structural domains functional ontology (SDFO). Briefly, the algorithm iteratively identifies GO terms closest to a predefined IC (say 1) as a seed until all paths have been searched in the DAG, on the condition that one and only one GO term can be identified per path. If multiple GO terms with identical IC are identified in the same path, we filter out those parental terms. Once a GO term is identified, all terms in the path in which that term is located will be marked for being immune from further search. The outputs are those identified GO terms with IC falling in the range (say, 1 ± 0.25). We run the algorithm using each of four seed ICs (i.e. 0.5, 1, 1.5 and 2) to create SDFO, respectively corresponding to GO terms with four levels (least informative, moderately informative, informative, highly informative).
| (5) |
where IC for a term GOi is defined as negative log-transformation of the frequency of observing domains annotated to that term.
Comparing with existing manual annotation
Although it is hard to systematically evaluate the accuracy of domain-centric GOAs without gold-standard benchmarks, it is feasible to make comparisons with independent high-confidence annotations related to a specific functional category. Since the procedures proposed above are not biased toward a specific function, such comparisons can give us an intuitive overview of performance. In this aspect, the DNA-binding domain (DBD) database (33), containing a manually curated list of sequence-specific DNA-binding domains at the SCOP superfamily level, can be of use. For this comparison, we treat the GO term ‘DNA binding’ [GO:0003677] as an equivalent functional annotation for the DNA-binding domains. Out of 38 domains in DBD, 24 can be found in at least one UniProt annotated term, and thus can be used for the comparisons in this study. As shown in Figure 2C, domains annotated to ‘DNA binding’, which are inferred using all UniProt sequences, highly overlap with those in DBD, taking up almost 75% accuracy (18/24). Moreover, there are up to 3.75-fold increases in coverage (90/24). Further inspection of those truly domain-centric annotations supported by both singleton domain sequences and all sequences, 8 out of 14 domains can be found in DBD. It indicates that at least half of DNA-binding domains function independently as DNA binding domains, regardless of the presence of other domains in multi-domain proteins. Of note, the GO term together with these 14 high-quality domain-centric annotations, are included in SDFO at the informative level. Since the GO term ‘DNA binding’ [GO:0003677] may cover non-specific binding, which is excluded from consideration in DBD, we also used other GO terms (such as those related to transcription regulation) for comparison. Indeed, similar results can be obtained when focusing on the GO term ‘transcription regulator activity’ [GO:0030528] (Figure 2D).
This demonstration partially validates the power of our procedure in developing domain-centric GOAs. First, it is a good starting point for creating a compatible ontology with protein-centric GO. Second, truly domain-centric functional annotations make it possible to study the extent of domain combinations on the neo-functions. Last but not least, most users may not care too much about whether annotations are truly domain-centric or not, so it would be very convenient to get high-coverage and high-quality lists of domains related to a specific functional categories of interest. Practically, due to limitations in the number of singleton domain proteins available for statistical testing, we must use all proteins (including multi-domain proteins) to perform the inference of phenotypic annotations below.
Extension to phenotypic annotations of structural domains
Like GO, phenotypic ontologies such as MPO and HPO have been developed to classify and organize phenotypic information related to the mouse and the human from the very general at the top to more specific terms in the DAG. MPO describes phenotypes of the mouse after a specific gene is genetically disrupted (27), while HPO captures phenotypic abnormalities that are described in OMIM, along with the corresponding disease-causing genes (28). Similar to statistical inference of domain-centric GOAs, we were also motivated to annotate structural domains with MPO (and HPO) that likely underlie the protein/gene-level phenotypic abnormalities. Similar procedures to those described in Figure 2A were applied except for two modifications. First, we only consider the longest transcript to ensure that the one-gene–one-protein mapping is valid, as these phenotypic annotations are gene orientated rather than protein based. Second, associations between domains and phenotypes are only supported by all proteins, due to the failure of statistical testing using insufficient number of singleton domain proteins in the mouse genome (or human genome). Figure 2E summarizes the total number of domains that can be annotated by MPO and HPO. This preliminary summary reveals that some structural domains can be of relevance to phenotypic studies. As the coverage and accuracy of these primary databases improve, we expect that much more domain-centric phenotypic annotations will be generated using the proposed procedures.
FUTURE DIRECTIONS
The most important goal we hope to achieve in the long term is to partially automate the model-building procedure, and in collaboration with SCOP and ASTRAL to move towards rolling weekly updates in place of the periodic updates. We plan to continue to develop SUPERFAMILY as a tool for understanding the evolution of protein domains and the genomes/metagenomes in which they are found.
FUNDING
Funding for open access charge: European Union Framework Program 7 Impact grant (grant number 213037); Biotechnology and Biological Sciences Research Council (grant number G022771).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank their user-base, in particular those users who have contributed to the development of SUPERFAMILY via their feedback with constructive criticisms or enthusiasm and by sharing with them the ways in which the resource has and can help in their research. They also thank Aare Abroi for advice regarding viral genomes and Matt Oates for schema documentation.
REFERENCES
- 1.Gough J, Chothia C. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 2002;30:268–272. doi: 10.1093/nar/30.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 2004;32:D235–D239. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;35:D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY––sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The UniProt Consortium. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 8.Karplus K, Barrett C, Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 9.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 10.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pethica R, Barker G, Kovacs T, Gough J. TreeVector: scalable, interactive, phylogenetic trees for the web. PLoS One. 2010;5:e8934. doi: 10.1371/journal.pone.0008934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rackham OJ, Madera M, Armstrong CT, Vincent TL, Woolfson DN, Gough J. The evolution and structure prediction of coiled coils across all genomes. J. Mol. Biol. 2010;403:480–493. doi: 10.1016/j.jmb.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 15.Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munroe DJ, Harris TJ. Third-generation sequencing fireworks at Marco Island. Nat. Biotechnol. 2010;28:426–428. doi: 10.1038/nbt0510-426. [DOI] [PubMed] [Google Scholar]
- 17.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP. Computational solutions to large-scale data management and analysis. Nat. Rev. Genet. 2010;11:647–657. doi: 10.1038/nrg2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madera M, Gough J. A comparison of profile hidden Markov model procedures for remote homology detection. Nucleic Acids Res. 2002;30:4321–4328. doi: 10.1093/nar/gkf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009–an integrated gene ontology annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velankar S, Best C, Beuth B, Boutselakis CH, Cobley N, Sousa Da Silva AW, Dimitropoulos D, Golovin A, Hirshberg M, John M, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2010;38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 23.Gough J. Genomic scale sub-family assignment of protein domains. Nucleic Acids Res. 2006;34:3625–3633. doi: 10.1093/nar/gkl484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forslund K, Sonnhammer EL. Predicting protein function from domain content. Bioinformatics. 2008;24:1681–1687. doi: 10.1093/bioinformatics/btn312. [DOI] [PubMed] [Google Scholar]
- 26.Lopez D, Pazos F. Gene ontology functional annotations at the structural domain level. Proteins. 2009;76:598–607. doi: 10.1002/prot.22373. [DOI] [PubMed] [Google Scholar]
- 27.Smith CL, Eppig JT. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers MF, Ben-Hur A. The use of gene ontology evidence codes in preventing classifier assessment bias. Bioinformatics. 2009;25:1173–1177. doi: 10.1093/bioinformatics/btp122. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate––a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 32.Alterovitz G, Xiang M, Mohan M, Ramoni MF. GO PaD: the gene ontology partition database. Nucleic Acids Res. 2007;35:D322–D327. doi: 10.1093/nar/gkl799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD––taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]