Abstract
The Gene Expression Database (GXD) is a community resource of mouse developmental expression information. GXD integrates different types of expression data at the transcript and protein level and captures expression information from many different mouse strains and mutants. GXD places these data in the larger biological context through integration with other Mouse Genome Informatics (MGI) resources and interconnections with many other databases. Web-based query forms support simple or complex searches that take advantage of all these integrated data. The data in GXD are obtained from the literature, from individual laboratories, and from large-scale data providers. All data are annotated and reviewed by GXD curators. Since the last report, the GXD data content has increased significantly, the interface and data displays have been improved, new querying capabilities were implemented, and links to other expression resources were added. GXD is available through the MGI web site (www.informatics.jax.org), or directly at www.informatics.jax.org/expression.shtml.
INTRODUCTION
As a primary mammalian model of human disease, the mouse is used extensively for expression studies to determine the role of genes that function in molecular pathways during developmental and disease processes. With a focus on endogenous gene expression during development, the Gene Expression Database (GXD) collects data from the scientific literature, from individual laboratories, and from large-scale data providers. It makes these data readily available to the research community in a highly curated and integrated format that allows for a large variety of database queries. GXD captures a broad spectrum of assay types, including RNA in situ hybridization, immunohistochemistry, knock-in reporter assays, northern blot, western blot, RT–PCR, RNase protection and S1 nuclease assays. It covers all developmental stages and tissues and includes data from many different mouse strains and mutants, giving researchers a tool to examine the effects of mutations on gene expression. GXD forms an important and integral component of the larger Mouse Genome Informatics (MGI) resource. Therefore, the expression data are fully integrated with mouse genetic, sequence, functional and phenotypic information (1–4). MGI maintains further links to many other resources such as GenBank, gene model resources, Entrez Gene, UniProt, InterPro, Online Mendelian Inheritance in Man (OMIM) and the International Knockout Mouse Consortium (IKMC) among others (5–14). This robust integration puts the expression data in GXD into a much larger biological and analytical context.
Other databases that store mouse expression information have been developed in recent years. They store data from one or two specific assay types and/or focus on specific developmental stages; they are often dedicated to specific data generation projects (15–22). As will be evident from this article, GXD is working with those resources that are complementary to GXD, adding value through data integration and the implementation of new interconnections. Due to its broad scope, its extensive data curation and integration efforts, and the resulting querying capabilities, GXD continues to provide a unique resource to the biomedical research community. GXD is updated daily. GXD and its query interfaces have been described earlier (23–27). Here, we report on our recent progress in terms of data acquisition, and on the implementation of new query and display features.
NEW GXD HOMEPAGE
To present the objectives of GXD more clearly and to make the database more intuitive to use, we redesigned the GXD homepage (www.informatics.jax.org/expression.shtml). The new layout provides clear access to the various query forms, with short descriptors for each form. The ‘Frequently Asked Questions (FAQ’s)’ section provides links to brief on-line tutorials demonstrating how one can search for different types of data in GXD. The ‘GXD Includes’ section provides information about the current data content in GXD, such as the number of genes with annotated expression data, the number of expression results and the number of images in the database. The ‘Gene Expression News’ section informs users when new features, capabilities and data sets become available. A series of tabs at the bottom of the home page provides access to help documentation and data policies, information about GXD and its collaborators, and links to guidelines and tools that help researchers to submit data electronically. That GXD is also an integral component of MGI is made transparent through the use of a central ‘Quick Search’ (see below), a common navigation bar and common drop-menus and tab choices that direct users to various data sets, search forms, tools and other resources. Large icons on the MGI homepage provide visual cues to the various core areas, including expression (GXD).
DATA CONTENT
Literature Summary
In the Literature Summary, GXD provides users with a way to quickly determine what mouse developmental expression data are available in the scientific literature. The staff of GXD searches the scientific literature for publications that present endogenous gene expression experiments during mouse development. In a first annotation step for each publication, the genes analyzed, the ages of mice used in the experiment, and the type of experimental assay performed for each gene are recorded and entered into the database. These data are easily searched using the ‘Gene Expression Literature Query Form’. These queries can also include citation (author, journal, year) and abstract information. However, this tool takes users further than a Pubmed search because the data in the GXD Literature Summary are based on the curation of the full-text of the paper, including supplemental information, and annotations are standardized with regard to gene, age and assay type information. The Literature Summary is comprehensive and up-to-date. It includes all journal articles containing expression data during mouse development from 1993 to the present and all articles from major developmental journals since 1990. Currently, the GXD Literature Summary has 108 604 records covering data for 13 619 genes from 17 521 references.
Gene expression data
Beyond summaries, GXD also provides detailed records of experimental expression results. GXD assay records contain the authors’ description of the tissue pattern and strength of expression, translated into standard terminology (see below), the probe or antibody information available, as well as the specimen age, genetic background and preparation (Figure 1). The expression information is recorded in standardized formats by making extensive use of controlled vocabularies and ontologies, thus enabling data integration and complex querying capabilities. To capture the author’s descriptions of where expression was or was not detected, GXD uses a dictionary of anatomical terms that lists the anatomical structures for each developmental stage in a hierarchical fashion. In this way, expression data from assays with differing spatial resolution can be recorded in a consistent manner, and the expression pattern information becomes accessible to hierarchical searches. The developmental part of the anatomical dictionary was established by our collaborators from the EMAGE project (28) and is being extended and refined jointly; the postnatal part was developed by the GXD project (29).
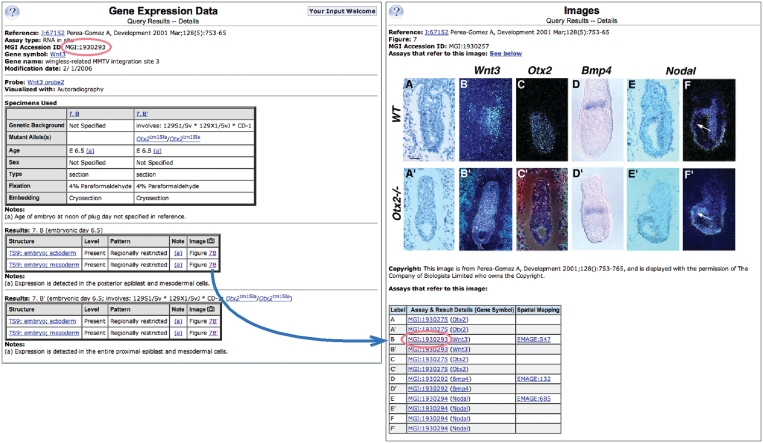
Figure 1.
Detailed gene expression data annotations and images. An assay record for Wnt3 (left) and the accompanying image page (right). The assay page includes: publication reference, gene symbol and name, link to probe/antibody information, specimen details, and the expression annotations for each specimen. Annotations include the anatomical structures in which expression was analyzed and the strength of expression in these structures. They refer to corresponding figures in publications or electronic submissions. Whenever possible, the images are included in GXD and assay records link to those images. The image page includes: publication reference, figure label, the figure, a caption and a copyright statement of the journal or the submitter. Figures from publications often include several image panes analyzing different genes and/or genetic backgrounds. The table below the figure indicates each pane and its corresponding assay record with links to that record, as illustrated by the red circle. For images that have been spatially mapped onto the Edinburgh Mouse Atlas, the table provides a link to the corresponding entry in EMAGE.
In addition to curating expression data from the literature on a daily basis, GXD continues to work with individual laboratories and large-scale data providers to bring their data sets into the database. Recently acquired large data sets include: an RT–PCR screen of more than 800 genes in pre-implantation embryos (30); an RNA in situ hybridization screen of 293 genes at E7.5 (31); an RNA in situ hybridization screen of more than 200 genes in the developing hypothalamus (32); a set of knock-in lacZ reporter studies from Deltagen Inc.; an RNA in situ hybridization data for 745 genes at E14.5 from GenePaint (22) and a data set from the Eurexpress project (www.eurexpress.org), covering RNA in situ hybridization data for 6409 genes at E14.5. In all these cases, GXD staff worked with the data providers to bring the data into a standardized format that could be incorporated into GXD and to resolve issues pertinent to nomenclature, incompleteness, and referential integrity. Upon computational and manual review, the data were bulk-loaded into GXD.
Due to all these efforts, the content of expression data in GXD has increased tremendously in recent years. The database currently holds almost 930 000 expression results from 45 305 expression assays for 12 139 genes. This includes expression data from 1503 different mouse mutants.
ACCESSING GXD’S EXPRESSION DATA
An effective way to access the expression information for a single gene is to look up the ‘Expression’ section on the corresponding gene detail page (Figure 2). Gene detail pages can be easily found via the ‘Quick Search’ tool placed at the top of all MGI pages. Because these pages are a central hub within MGI, we have improved the layout of the expression section, implemented a new images summary (see below), and added new gene-specific links to external expression resources, including the Allen Brain Atlas (18), GENSAT (17), GEO (33) and ArrayExpress (34). Future work will include the implementation of links to model organisms from other species that hold developmental expression data, such as ZFIN (35), GEISHA (36), Xenbase (37) and Flybase (38).
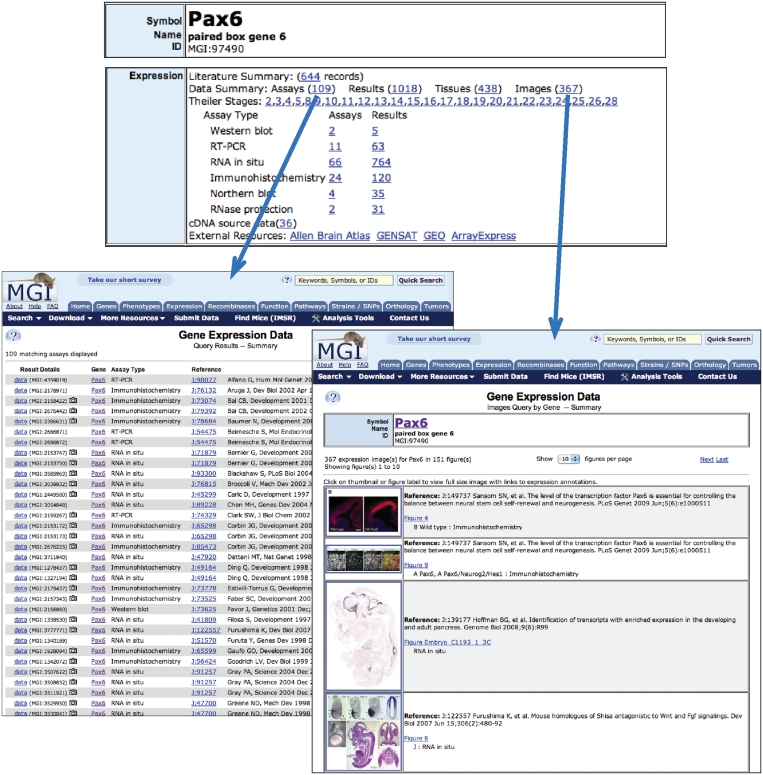
Figure 2.
Expression data summaries. Gene detail pages in MGI feature an ‘Expression’ section that summarizes the GXD data available for the gene from a variety of perspectives: literature content, assays, results, tissues, images, developmental stages and assay types (top). Links to the respective summaries provide quick access to the actual data. Gene-specific links to the Allen Brain Atlas, GENSAT, GEO and ArrayExpress are provided as well. Assay summaries (bottom left) link to the detailed assay records (‘data’) and show camera icons to indicate assay records with image links. The image summary (bottom right) lists thumbnails of all expression images for a gene and links to the full-size image page.
The query forms available from the GXD homepage, as well as from the MGI search menu, provide more direct and more powerful means to access expression data. The ‘Gene Expression Data Query Form’ includes search fields that allow users to specify genes, tissue, developmental stage and expression assay type, as well as genome coordinates and gene ontology (GO) terms. Users can search for instances where expression was detected or not detected, as well as for expression data in specific mutants. Thus, this form enables both simple and very complex queries. The ‘Expanded Expression Data Query Form’ allows users to find genes that are expressed in one set of tissues and/or developmental stages, but not in others. The ‘Mouse Anatomical Dictionary Browser’ lets users search or browse for specific anatomical structures and the expression data associated with them. The ‘EMAGE Anatomical Section Browser’ uses the Edinburgh Mouse Atlas as a starting point for identifying anatomical structures and expression data associated with them in GXD and EMAGE.
Searches from the query form as well as the links from the expression section on the gene detail page lead to data summaries listing all assays/results that match the user’s criteria. We have improved the layout of these summaries. They now feature clear ‘data’ links that take the user to the detailed entries, as well as camera icons that indicate if the detailed entries contain links to primary image data (Figures 1 and 2).
Other avenues in MGI can be used to access the expression data as well. We have increased the utility of the Quick Search tool by adding the capability to return expression results if the user enters anatomical terms. We have expanded the Batch Query tool so that it can return GXD expression data for a list of genes or sequence IDs, as well as export expression data as Excel worksheets or tab-delimited text files (Figure 3).
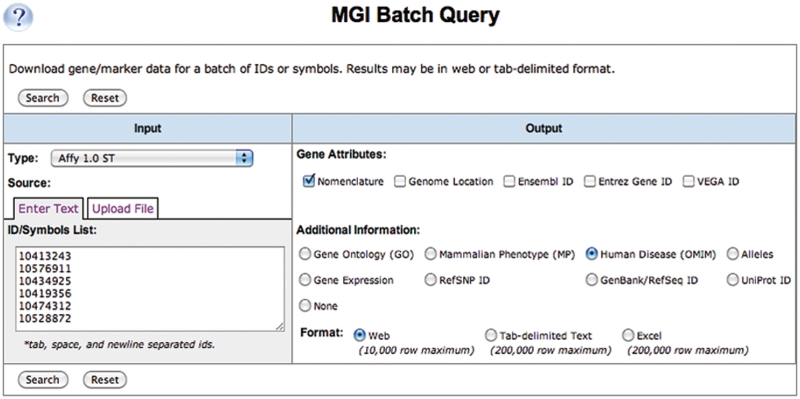
Figure 3.
MGI Batch Query. Batch queries can return GXD expression data for a list of genes or sequence IDs. Affymetrix probe set IDs are also accepted as input. Because MGI maintains up-to-date mappings of microarray probe set-to-gene associations, users can easily obtain MGI information about genes for specific sets of microarray probes, such as expression, phenotypic or, as shown, human disease data. Query results are available as web page displays, as tab-delimited text files or as Excel files. The Batch Query tool can be accessed at http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=batchQF.
In addition to implementing gene specific links to the microarray gene expression data at GEO and ArrayExpress, we have, in collaboration with the Mouse Genome Database (MGD) project, developed new tools that allow researchers to combine microarray expression results with data in MGI. Specifically, we have begun to provide up-to-date mappings between microarray probe sets and genes so that users can query MGI by microarray probe set ID to retrieve the associated gene, or download reports of all probe set-to-gene mappings. We have also enhanced the MGI Batch Query tool so that it accepts sets of microarray probe IDs. Therefore, one can easily extract information about genes for sets of microarray probes, including RNA in situ hybridization, immunohistochemistry and RT–PCR expression data, GO annotations, mouse phenotype data and disease data associated with the orthologous human genes. Query results are available as web page displays and can also be downloaded as tab-delimited text and thus fed into other analysis tools (Figure 3).
GXD is implemented in the Sybase relational database management system. If a user’s queries are beyond the capabilities of our web-based forms, custom SQLs or direct SQL access can be requested via our user support group. In addition, a number of reports for specific data sets, such as a list of all the genes that have annotated expression data, are produced nightly and are available on the website.
IMAGES IN GXD
Whenever possible, the standardized text annotations of expression data are linked to the primary image data. There are currently more than 191 099 images in GXD. Images are accessible via the ‘Expression’ section on the gene detail page. The recently implemented image summary displays all the images for the corresponding gene as thumbnails (Figure 2). These thumbnails are then linked to the full-sized image page. However, users are not restricted to selecting images on a per gene basis. Because all images are extensively indexed via the text annotations, they are accessible through the different search parameters that GXD provides on its query forms. Query result summaries display camera icons for entries that contain image data. The detailed entries then link directly to primary images obtained from the publication or from the data provider.
As part of our longstanding collaboration with the EMAGE project, which aims to ultimately create a resource that fully combines standardized text based and graphical means for storing and querying expression data (39), GXD makes all its in situ images and pertinent text annotations available to EMAGE. EMAGE then provides 3D spatial mappings of selected in situ data. The image detail pages in GXD now link to entries in EMAGE that show the image spatially mapped onto the 3D atlas (Figure 1).
Large-scale in situ hybridization projects have contributed both data and images to GXD. Two of them, GenePaint and Eurexpress, provide additional images and features for image viewing at their web sites. Therefore, GXD has added links from the image page at GXD to the corresponding image entries at GenePaint and Eurexpress. These successful collaborations illustrate how community resources such as GXD and project-oriented resources such as GenePaint and Eurexpress can work with each other. GXD adds value to the incorporated data by integrating them with other expression data and with all the other types of data stored in MGI and by providing unique and powerful querying capabilities. Further, GXD maintains the data and keeps them current with regard to nomenclature and data connections. Conversely, the GenePaint and Eurexpress data significantly enrich GXD, and the links implemented to GenePaint and Eurexpress provide additional utility such as high-resolution images and zoom functions.
DIRECT ELECTRONIC CONTRIBUTION OF DATA
Users can contribute data directly to GXD. Guidelines for electronic submission are available on the web site, and a GXD staff member is available to assist. GXD has developed an Excel-based program, the Gene Expression Notebook (GEN), which provides users with a template for submitting expression data to GXD. A variety of other formats are also accepted. The GEN is also useful in the lab for maintaining expression data and is available for free at http://www.informatics.jax.org/mgihome/GXD/GEN/index.shtml. All data submitted to GXD are given accession IDs that can be referenced in publications and grant applications.
USER SUPPORT
GXD provides users with extensive on-line documentation available by clicking on the question marks available on many pages. In addition, user support staff can be contacted via the ‘Help’ or ‘Contact Us’ links on the website, by email (mgi-help@informatics.jax.org) or by telephone (207-288-6445).
CITING GXD
The following citation format is suggested when referring to data from GXD: These data were retrieved from the GXD, and MGI, The Jackson Laboratory, Bar Harbor, Maine, USA (URL: http://www.informatics.jax.org). [Type in date (month, year) when you retrieved the data cited.] To reference the database itself, please cite this article.
FUNDING
National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development grant HD033745. Funding for open access charge: National Institutes of Health grant HD033745.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank our colleagues from the other MGI projects for their contributions to the GXD project and to the larger integrated MGI resource.
REFERENCES
- 1.Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT the Mouse Genome Database Group. The mouse genome database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley DA, Krupke DM, Vincent MJ, Sundberg JP, Bult CJ, Eppig JT. Mouse tumor biology database (MTB): status update and future directions. Nucleic Acids Res. 2007;35:D638–D642. doi: 10.1093/nar/gkl983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evsikov AV, Dolan ME, Genrich MP, Patek E, Bult CJ. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfsberg TG. Using the NCBI map viewer to browse genomic sequence data. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi0105s29. Chapter 1, Unit 1.5.1–1.5.25. [DOI] [PubMed] [Google Scholar]
- 10.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Mouse Knockout Consortium. Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ, et al. An anatomy of normal and malignant gene expression. Proc. Natl Acad. Sci. USA. 2002;99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Divina P, Forejt J. The Mouse SAGE Site: database of public mouse SAGE libraries. Nucleic Acids Res. 2004;32:D482–D483. doi: 10.1093/nar/gkh058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geschwind D. GENSAT: a genomic resource for neuroscience research. Lancet Neurol. 2004;3:82. doi: 10.1016/s1474-4422(03)00658-6. [DOI] [PubMed] [Google Scholar]
- 18.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 19.Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL the GUDMAP project. GUDMAP: the genitourinary developmental molecular anatomy project. J. Am. Soc. Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 21.Richardson L, Venkataraman S, Stevenson P, Yang Y, Burton N, Rao J, Fisher M, Baldock RA, Davidson DR, Christiansen JH. EMAGE mouse embryo spatial gene expression database: 2010 update. Nucleic Acids Res. 2010;38:D703–D709. doi: 10.1093/nar/gkp763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2007 update. Nucleic Acids Res. 2007;35:D618–D623. doi: 10.1093/nar/gkl1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill DP, Begley DA, Finger JH, Hayamizu TF, McCright IJ, Smith CM, Beal JS, Corbani LE, Blake JA, Eppig JT, et al. The mouse gene expression database (GXD): updates and enhancements. Nucleic Acids Res. 2004;32:D568–D571. doi: 10.1093/nar/gkh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringwald M, Eppig JT, Begley DA, Corradi JP, McCright IJ, Hayamizu TF, Hill DP, Kadin JA, Richardson JE. The Mouse Gene Expression Database (GXD) Nucleic Acids Res. 2001;29:98–101. doi: 10.1093/nar/29.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ringwald M, Eppig JT, Kadin JA, Richardson JE. GXD: a gene expression database for the laboratory mouse: current status and recent enhancements. The Gene Expression Database group. Nucleic Acids Res. 2000;28:115–119. doi: 10.1093/nar/28.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringwald M, Mangan ME, Eppig JT, Kadin JA, Richardson JE. GXD: a gene expression database for the laboratory mouse. The Gene Expression Database Group. Nucleic Acids Res. 1999;27:106–112. doi: 10.1093/nar/27.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev. 1998;74:111–120. doi: 10.1016/s0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 29.Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The adult mouse anatomical dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo G, Huss M, Tong GQ, Wang C, LiSun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Tamplin OJ, Kinzel D, Cox BJ, Bell CE, Rossant J, Lickert H. Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC Genomics. 2008;9:511. doi: 10.1186/1471-2164-9-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, et al. A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update––from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramachandran S, Ruef B, Pich C, Sprague J. Exploring Zebrafish genomic, functional and phenotypic data using ZFIN. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi0118s31. Chapter 1, Unit 1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darnell DK, Kaur S, Stanislaw S, Davey S, Konieczka JH, Yatskievych TA, Antin PB. GEISHA: an in situ hybridization gene expression resource for the chicken embryo. Cytogenet. Genome Res. 2007;117:30–35. doi: 10.1159/000103162. [DOI] [PubMed] [Google Scholar]
- 37.Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2010;38:D607–D612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drysdale R the FlyBase Consortium. FlyBase: a database for the Drosophila research community. Methods Mol. Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- 39.Ringwald M, Baldock R, Bard J, Kaufman M, Eppig JT, Richardson JE, Nadeau JH, Davidson D. A database for mouse development. Science. 1994;265:2033–2034. doi: 10.1126/science.8091224. [DOI] [PubMed] [Google Scholar]